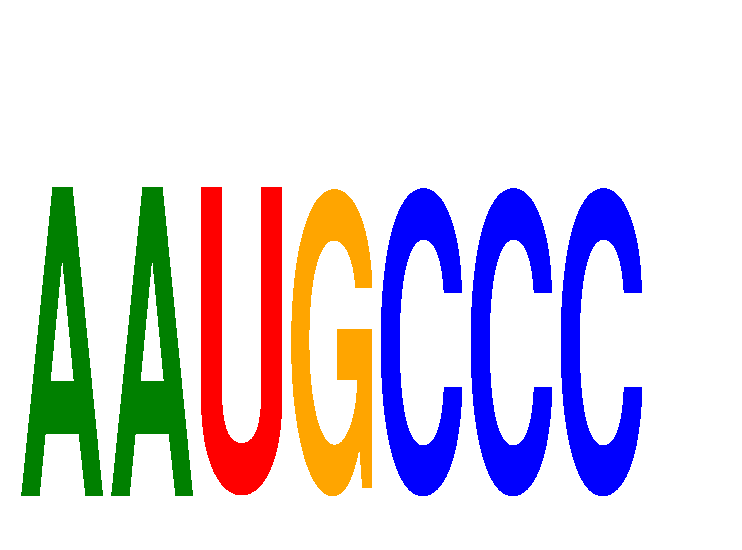Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.06
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-365a-3p
|
MIMAT0000710 |
|
hsa-miR-365b-3p
|
MIMAT0022834 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_68165657 | 0.07 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr12_+_72666407 | 0.07 |
ENST00000261180.4 |
TRHDE |
thyrotropin-releasing hormone degrading enzyme |
| chr13_-_21476900 | 0.06 |
ENST00000400602.2 ENST00000255305.6 |
XPO4 |
exportin 4 |
| chr15_+_52121822 | 0.05 |
ENST00000558455.1 ENST00000308580.7 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr1_-_153588334 | 0.04 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr22_+_37309662 | 0.04 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_-_208634287 | 0.04 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chr3_+_113775576 | 0.04 |
ENST00000485050.1 ENST00000281273.4 |
QTRTD1 |
queuine tRNA-ribosyltransferase domain containing 1 |
| chr3_-_123603137 | 0.04 |
ENST00000360304.3 ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK |
myosin light chain kinase |
| chr5_-_39074479 | 0.04 |
ENST00000514735.1 ENST00000296782.5 ENST00000357387.3 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
| chr1_-_23857698 | 0.03 |
ENST00000361729.2 |
E2F2 |
E2F transcription factor 2 |
| chr4_-_149365827 | 0.03 |
ENST00000344721.4 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr9_-_23821273 | 0.03 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr10_-_62149433 | 0.03 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr11_+_34642656 | 0.03 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr11_-_102962929 | 0.03 |
ENST00000260247.5 |
DCUN1D5 |
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr3_+_14989076 | 0.03 |
ENST00000413118.1 ENST00000425241.1 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr3_+_150804676 | 0.03 |
ENST00000474524.1 ENST00000273432.4 |
MED12L |
mediator complex subunit 12-like |
| chr8_+_28351707 | 0.03 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr10_-_105615164 | 0.03 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr9_-_14314066 | 0.03 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr7_+_97736197 | 0.02 |
ENST00000297293.5 |
LMTK2 |
lemur tyrosine kinase 2 |
| chr19_+_33166313 | 0.02 |
ENST00000334176.3 |
RGS9BP |
regulator of G protein signaling 9 binding protein |
| chr15_+_75074385 | 0.02 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr5_+_149109825 | 0.02 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr14_-_99737565 | 0.02 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr5_-_43313574 | 0.02 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr17_-_48072574 | 0.02 |
ENST00000434704.2 |
DLX3 |
distal-less homeobox 3 |
| chr8_-_141645645 | 0.02 |
ENST00000519980.1 ENST00000220592.5 |
AGO2 |
argonaute RISC catalytic component 2 |
| chr8_-_57906362 | 0.02 |
ENST00000262644.4 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
| chr7_-_83824169 | 0.02 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_+_63606373 | 0.02 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr2_-_153574480 | 0.02 |
ENST00000410080.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr12_-_42538657 | 0.02 |
ENST00000398675.3 |
GXYLT1 |
glucoside xylosyltransferase 1 |
| chr1_-_244013384 | 0.02 |
ENST00000366539.1 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chrX_+_69664706 | 0.02 |
ENST00000194900.4 ENST00000374360.3 |
DLG3 |
discs, large homolog 3 (Drosophila) |
| chr9_+_130548297 | 0.02 |
ENST00000373264.4 |
CDK9 |
cyclin-dependent kinase 9 |
| chr8_+_124780672 | 0.02 |
ENST00000521166.1 ENST00000334705.7 |
FAM91A1 |
family with sequence similarity 91, member A1 |
| chr3_-_47823298 | 0.02 |
ENST00000254480.5 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr4_-_11431389 | 0.02 |
ENST00000002596.5 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr19_+_50094866 | 0.01 |
ENST00000418929.2 |
PRR12 |
proline rich 12 |
| chr13_-_46425865 | 0.01 |
ENST00000400405.2 |
SIAH3 |
siah E3 ubiquitin protein ligase family member 3 |
| chr11_+_128563652 | 0.01 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_-_113465065 | 0.01 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_+_68042495 | 0.01 |
ENST00000344096.3 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr3_+_23986748 | 0.01 |
ENST00000312521.4 |
NR1D2 |
nuclear receptor subfamily 1, group D, member 2 |
| chr12_+_5019061 | 0.01 |
ENST00000382545.3 |
KCNA1 |
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr1_-_85462623 | 0.01 |
ENST00000370608.3 |
MCOLN2 |
mucolipin 2 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr4_-_74124502 | 0.01 |
ENST00000358602.4 ENST00000330838.6 ENST00000561029.1 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr12_-_67072714 | 0.01 |
ENST00000545666.1 ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr8_+_1449532 | 0.01 |
ENST00000421627.2 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
| chr6_-_88411911 | 0.01 |
ENST00000257787.5 |
AKIRIN2 |
akirin 2 |
| chr3_+_51575596 | 0.01 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr19_+_56186557 | 0.01 |
ENST00000270460.6 |
EPN1 |
epsin 1 |
| chr7_-_111846435 | 0.01 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr7_+_4721885 | 0.01 |
ENST00000328914.4 |
FOXK1 |
forkhead box K1 |
| chr18_+_43753974 | 0.01 |
ENST00000282059.6 ENST00000321319.6 |
C18orf25 |
chromosome 18 open reading frame 25 |
| chr10_-_103874692 | 0.01 |
ENST00000361198.5 |
LDB1 |
LIM domain binding 1 |
| chr2_-_64881018 | 0.01 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr12_-_31479045 | 0.01 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr9_-_117880477 | 0.01 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr7_+_94285637 | 0.01 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr14_-_36989427 | 0.01 |
ENST00000354822.5 |
NKX2-1 |
NK2 homeobox 1 |
| chr22_+_32340481 | 0.01 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr22_+_21271714 | 0.01 |
ENST00000354336.3 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr2_-_39348137 | 0.01 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr5_-_168006591 | 0.01 |
ENST00000239231.6 |
PANK3 |
pantothenate kinase 3 |
| chr4_+_38665810 | 0.01 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr9_+_128024067 | 0.01 |
ENST00000461379.1 ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1 |
GTPase activating protein and VPS9 domains 1 |
| chr14_-_21905395 | 0.01 |
ENST00000430710.3 ENST00000553283.1 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
| chr11_-_27528301 | 0.01 |
ENST00000524596.1 ENST00000278193.2 |
LIN7C |
lin-7 homolog C (C. elegans) |
| chr1_-_28415204 | 0.01 |
ENST00000373871.3 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr16_+_70557685 | 0.01 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr12_-_49449107 | 0.01 |
ENST00000301067.7 |
KMT2D |
lysine (K)-specific methyltransferase 2D |
| chr6_-_3457256 | 0.01 |
ENST00000436008.2 |
SLC22A23 |
solute carrier family 22, member 23 |
| chr1_-_156051789 | 0.01 |
ENST00000532414.2 |
MEX3A |
mex-3 RNA binding family member A |
| chr17_-_40273348 | 0.01 |
ENST00000225916.5 |
KAT2A |
K(lysine) acetyltransferase 2A |
| chr7_-_151217001 | 0.01 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr4_+_91048706 | 0.01 |
ENST00000509176.1 |
CCSER1 |
coiled-coil serine-rich protein 1 |
| chr17_-_27621125 | 0.01 |
ENST00000579665.1 ENST00000225388.4 |
NUFIP2 |
nuclear fragile X mental retardation protein interacting protein 2 |
| chr8_+_128426535 | 0.01 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr17_-_19771216 | 0.01 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr3_-_79068594 | 0.01 |
ENST00000436010.2 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr17_-_16118835 | 0.01 |
ENST00000582357.1 ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1 |
nuclear receptor corepressor 1 |
| chrX_+_77359671 | 0.01 |
ENST00000373316.4 |
PGK1 |
phosphoglycerate kinase 1 |
| chr10_+_101419187 | 0.01 |
ENST00000370489.4 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr19_+_46850251 | 0.01 |
ENST00000012443.4 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr1_+_2985760 | 0.01 |
ENST00000378391.2 ENST00000514189.1 ENST00000270722.5 |
PRDM16 |
PR domain containing 16 |
| chr6_+_106546808 | 0.01 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr17_+_36508111 | 0.01 |
ENST00000331159.5 ENST00000577233.1 |
SOCS7 |
suppressor of cytokine signaling 7 |
| chr6_+_34857019 | 0.01 |
ENST00000360359.3 ENST00000535627.1 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr1_-_35658736 | 0.01 |
ENST00000357214.5 |
SFPQ |
splicing factor proline/glutamine-rich |
| chr22_-_42017021 | 0.01 |
ENST00000263256.6 |
DESI1 |
desumoylating isopeptidase 1 |
| chr16_+_50582222 | 0.01 |
ENST00000268459.3 |
NKD1 |
naked cuticle homolog 1 (Drosophila) |
| chr16_+_67063036 | 0.01 |
ENST00000290858.6 ENST00000564034.1 |
CBFB |
core-binding factor, beta subunit |
| chr2_-_183903133 | 0.01 |
ENST00000361354.4 |
NCKAP1 |
NCK-associated protein 1 |
| chr3_-_48885228 | 0.01 |
ENST00000454963.1 ENST00000296446.8 ENST00000419216.1 ENST00000265563.8 |
PRKAR2A |
protein kinase, cAMP-dependent, regulatory, type II, alpha |
| chr12_-_82153087 | 0.01 |
ENST00000547623.1 ENST00000549396.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_-_3631635 | 0.00 |
ENST00000262848.5 |
PRKX |
protein kinase, X-linked |
| chr17_+_4613918 | 0.00 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chrX_-_74376108 | 0.00 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr9_+_116638562 | 0.00 |
ENST00000374126.5 ENST00000288466.7 |
ZNF618 |
zinc finger protein 618 |
| chr20_-_1974692 | 0.00 |
ENST00000217305.2 ENST00000539905.1 |
PDYN |
prodynorphin |
| chr1_-_91487013 | 0.00 |
ENST00000347275.5 ENST00000370440.1 |
ZNF644 |
zinc finger protein 644 |
| chr8_+_54764346 | 0.00 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr5_+_74632993 | 0.00 |
ENST00000287936.4 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr5_+_148724993 | 0.00 |
ENST00000513661.1 ENST00000329271.3 ENST00000416916.2 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
| chr2_+_155554797 | 0.00 |
ENST00000295101.2 |
KCNJ3 |
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr15_+_93015151 | 0.00 |
ENST00000556865.1 |
C15orf32 |
chromosome 15 open reading frame 32 |
| chr2_+_134877740 | 0.00 |
ENST00000409645.1 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr1_-_205719295 | 0.00 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr10_-_735553 | 0.00 |
ENST00000280886.6 ENST00000423550.1 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr8_-_4852218 | 0.00 |
ENST00000400186.3 ENST00000602723.1 |
CSMD1 |
CUB and Sushi multiple domains 1 |
| chr7_-_141401951 | 0.00 |
ENST00000536163.1 |
KIAA1147 |
KIAA1147 |
| chr9_+_131445928 | 0.00 |
ENST00000372692.4 |
SET |
SET nuclear oncogene |
| chr22_+_42229100 | 0.00 |
ENST00000361204.4 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
| chr5_-_32444828 | 0.00 |
ENST00000265069.8 |
ZFR |
zinc finger RNA binding protein |
| chrX_+_73641286 | 0.00 |
ENST00000587091.1 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr7_-_32931387 | 0.00 |
ENST00000304056.4 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
| chr20_+_60697480 | 0.00 |
ENST00000370915.1 ENST00000253001.4 ENST00000400318.2 ENST00000279068.6 ENST00000279069.7 |
LSM14B |
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr11_-_102714534 | 0.00 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chrX_+_23352133 | 0.00 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr2_+_179345173 | 0.00 |
ENST00000234453.5 |
PLEKHA3 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr3_+_127872265 | 0.00 |
ENST00000254730.6 ENST00000483457.1 |
EEFSEC |
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr10_-_120514720 | 0.00 |
ENST00000369151.3 ENST00000340214.4 |
CACUL1 |
CDK2-associated, cullin domain 1 |
| chr7_-_27205136 | 0.00 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr12_+_2162447 | 0.00 |
ENST00000335762.5 ENST00000399655.1 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr8_-_38239732 | 0.00 |
ENST00000534155.1 ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr3_+_152552685 | 0.00 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chr9_-_127905736 | 0.00 |
ENST00000336505.6 ENST00000373549.4 |
SCAI |
suppressor of cancer cell invasion |
| chr9_+_114659046 | 0.00 |
ENST00000374279.3 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
| chr3_-_125094093 | 0.00 |
ENST00000484491.1 ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148 |
zinc finger protein 148 |
| chr16_-_67840442 | 0.00 |
ENST00000536251.1 ENST00000448631.2 ENST00000602677.1 ENST00000411657.2 ENST00000425512.2 ENST00000317506.3 |
RANBP10 |
RAN binding protein 10 |
| chr7_+_139044621 | 0.00 |
ENST00000354926.4 |
C7orf55-LUC7L2 |
C7orf55-LUC7L2 readthrough |
| chr5_-_100238956 | 0.00 |
ENST00000231461.5 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chrX_-_109561294 | 0.00 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr12_+_56137064 | 0.00 |
ENST00000257868.5 ENST00000546799.1 |
GDF11 |
growth differentiation factor 11 |
| chr15_-_64995399 | 0.00 |
ENST00000559753.1 ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2 |
ornithine decarboxylase antizyme 2 |
| chr2_+_131862900 | 0.00 |
ENST00000438882.2 ENST00000538982.1 ENST00000404460.1 |
PLEKHB2 |
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr9_-_136933134 | 0.00 |
ENST00000303407.7 |
BRD3 |
bromodomain containing 3 |
| chr12_-_75905374 | 0.00 |
ENST00000438169.2 ENST00000229214.4 |
KRR1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr2_+_46524537 | 0.00 |
ENST00000263734.3 |
EPAS1 |
endothelial PAS domain protein 1 |
| chr7_+_6414128 | 0.00 |
ENST00000348035.4 ENST00000356142.4 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| chr2_+_192542850 | 0.00 |
ENST00000410026.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr1_-_39339777 | 0.00 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr1_-_154600421 | 0.00 |
ENST00000368471.3 ENST00000292205.5 |
ADAR |
adenosine deaminase, RNA-specific |
| chr6_+_30294612 | 0.00 |
ENST00000440271.1 ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39 |
tripartite motif containing 39 |
| chr2_-_43453734 | 0.00 |
ENST00000282388.3 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
| chr10_-_120101804 | 0.00 |
ENST00000369183.4 ENST00000369172.4 |
FAM204A |
family with sequence similarity 204, member A |
| chr2_+_61108650 | 0.00 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr21_+_38071430 | 0.00 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr1_-_38325256 | 0.00 |
ENST00000373036.4 |
MTF1 |
metal-regulatory transcription factor 1 |
| chr18_+_32073253 | 0.00 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr11_-_77532050 | 0.00 |
ENST00000308488.6 |
RSF1 |
remodeling and spacing factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |