Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for ARID5A
Z-value: 0.54
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | ARID5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97203082_97203159, hg19_v2_chr2_+_97202480_97202499 | 0.11 | 8.0e-01 | Click! |
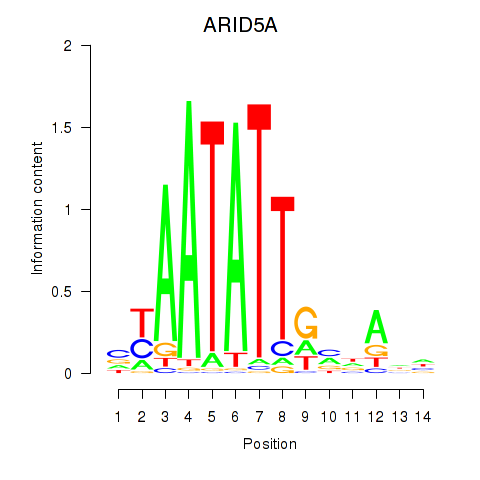
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602771 | 1.07 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr12_+_21207503 | 0.80 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr18_+_61554932 | 0.77 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr5_+_140220769 | 0.63 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr5_+_140165876 | 0.58 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr1_+_77333117 | 0.55 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_+_100737954 | 0.54 |
ENST00000296414.7 ENST00000512369.1 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr1_+_62439037 | 0.53 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr5_+_140261703 | 0.46 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr10_+_24755416 | 0.45 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr11_-_112034803 | 0.33 |
ENST00000528832.1 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_108408895 | 0.33 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chr10_+_47746929 | 0.32 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr6_+_33048222 | 0.31 |
ENST00000428835.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_85358850 | 0.31 |
ENST00000370611.3 |
LPAR3 |
lysophosphatidic acid receptor 3 |
| chr6_-_33048483 | 0.31 |
ENST00000419277.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr4_-_90756769 | 0.30 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_125648690 | 0.28 |
ENST00000436890.2 ENST00000358524.3 |
PATE2 |
prostate and testis expressed 2 |
| chr18_+_55816546 | 0.28 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_-_29343068 | 0.28 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr7_-_22233442 | 0.28 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_+_33043703 | 0.25 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr1_+_15256230 | 0.24 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr18_-_53089723 | 0.22 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr2_+_102618428 | 0.22 |
ENST00000457817.1 |
IL1R2 |
interleukin 1 receptor, type II |
| chr18_-_53069419 | 0.21 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr1_-_153013588 | 0.20 |
ENST00000360379.3 |
SPRR2D |
small proline-rich protein 2D |
| chrX_+_135614293 | 0.19 |
ENST00000370634.3 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr7_-_32111009 | 0.19 |
ENST00000396184.3 ENST00000396189.2 ENST00000321453.7 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chrX_-_32173579 | 0.19 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr9_-_140196703 | 0.19 |
ENST00000356628.2 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
| chr19_-_44174330 | 0.18 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr3_+_142342228 | 0.18 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr8_-_87081926 | 0.18 |
ENST00000276616.2 |
PSKH2 |
protein serine kinase H2 |
| chr3_+_113616317 | 0.18 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr11_-_5323226 | 0.17 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr14_+_22966642 | 0.17 |
ENST00000390496.1 |
TRAJ41 |
T cell receptor alpha joining 41 |
| chr19_-_44174305 | 0.17 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr17_+_55173933 | 0.16 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr1_-_113253977 | 0.16 |
ENST00000606505.1 ENST00000605933.1 |
RP11-426L16.10 |
Rho-related GTP-binding protein RhoC |
| chr5_+_147582348 | 0.16 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr18_+_28956740 | 0.16 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr6_+_24357131 | 0.16 |
ENST00000274766.1 |
KAAG1 |
kidney associated antigen 1 |
| chr1_+_81771806 | 0.16 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr1_+_13910194 | 0.16 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr2_+_102953608 | 0.15 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr18_-_61329118 | 0.15 |
ENST00000332821.8 ENST00000283752.5 |
SERPINB3 |
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr18_+_61223393 | 0.15 |
ENST00000269491.1 ENST00000382768.1 |
SERPINB12 |
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr5_+_140235469 | 0.14 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr8_-_124749609 | 0.14 |
ENST00000262219.6 ENST00000419625.1 |
ANXA13 |
annexin A13 |
| chr18_-_53253112 | 0.14 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr7_+_116593292 | 0.13 |
ENST00000393446.2 ENST00000265437.5 ENST00000393451.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr16_-_58328923 | 0.13 |
ENST00000567164.1 ENST00000219301.4 ENST00000569727.1 |
PRSS54 |
protease, serine, 54 |
| chr3_+_190333097 | 0.13 |
ENST00000412080.1 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chrX_+_107288239 | 0.13 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr13_+_43355732 | 0.13 |
ENST00000313851.1 |
FAM216B |
family with sequence similarity 216, member B |
| chr9_-_117853297 | 0.13 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr2_-_69180083 | 0.13 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr6_+_150920999 | 0.12 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_216978709 | 0.12 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr11_+_60197069 | 0.12 |
ENST00000528905.1 ENST00000528093.1 |
MS4A5 |
membrane-spanning 4-domains, subfamily A, member 5 |
| chrX_+_79675965 | 0.12 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chr10_+_90424196 | 0.12 |
ENST00000394375.3 ENST00000608620.1 ENST00000238983.4 ENST00000355843.2 |
LIPF |
lipase, gastric |
| chr5_-_87516448 | 0.12 |
ENST00000511218.1 |
TMEM161B |
transmembrane protein 161B |
| chr16_-_58328870 | 0.12 |
ENST00000543437.1 |
PRSS54 |
protease, serine, 54 |
| chr5_+_147582387 | 0.12 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr7_-_142120321 | 0.12 |
ENST00000390377.1 |
TRBV7-7 |
T cell receptor beta variable 7-7 |
| chr3_-_108672742 | 0.12 |
ENST00000261047.3 |
GUCA1C |
guanylate cyclase activator 1C |
| chr10_+_106113515 | 0.12 |
ENST00000369704.3 ENST00000312902.5 |
CCDC147 |
coiled-coil domain containing 147 |
| chr8_-_86253888 | 0.11 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr9_+_117904097 | 0.11 |
ENST00000374016.1 |
DEC1 |
deleted in esophageal cancer 1 |
| chr15_-_41522889 | 0.11 |
ENST00000458580.2 ENST00000314992.5 ENST00000558396.1 |
EXD1 |
exonuclease 3'-5' domain containing 1 |
| chr10_+_127661942 | 0.11 |
ENST00000417114.1 ENST00000445510.1 ENST00000368691.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chrX_-_100662881 | 0.11 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr8_-_66701319 | 0.11 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr13_+_43355683 | 0.11 |
ENST00000537894.1 |
FAM216B |
family with sequence similarity 216, member B |
| chr1_+_67632083 | 0.11 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chrX_-_74376108 | 0.10 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr1_-_203320617 | 0.10 |
ENST00000354955.4 |
FMOD |
fibromodulin |
| chr1_+_171154347 | 0.10 |
ENST00000209929.7 ENST00000441535.1 |
FMO2 |
flavin containing monooxygenase 2 (non-functional) |
| chr1_+_196743912 | 0.10 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr14_-_67878917 | 0.10 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr14_+_22670455 | 0.10 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr6_+_123317116 | 0.10 |
ENST00000275162.5 |
CLVS2 |
clavesin 2 |
| chr4_-_71532339 | 0.10 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr11_-_114466477 | 0.10 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr2_-_105030466 | 0.10 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr11_-_114466471 | 0.10 |
ENST00000424261.2 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr3_+_97868170 | 0.09 |
ENST00000437310.1 |
OR5H14 |
olfactory receptor, family 5, subfamily H, member 14 |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_+_26087509 | 0.09 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr6_+_26087646 | 0.09 |
ENST00000309234.6 |
HFE |
hemochromatosis |
| chr4_-_143227088 | 0.09 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr11_-_59950486 | 0.09 |
ENST00000426738.2 ENST00000533023.1 ENST00000420732.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr12_-_50236907 | 0.09 |
ENST00000333924.4 |
BCDIN3D |
BCDIN3 domain containing |
| chr19_-_42947121 | 0.09 |
ENST00000601181.1 |
CXCL17 |
chemokine (C-X-C motif) ligand 17 |
| chr4_-_89978299 | 0.09 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr16_-_29910365 | 0.09 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_158815588 | 0.09 |
ENST00000438394.1 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr20_-_1309809 | 0.09 |
ENST00000360779.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr11_+_128563652 | 0.09 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_107288197 | 0.09 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr5_+_54398463 | 0.09 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr6_-_10415218 | 0.09 |
ENST00000466073.1 ENST00000498450.1 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr11_+_55029628 | 0.08 |
ENST00000417545.2 |
TRIM48 |
tripartite motif containing 48 |
| chr8_+_53850991 | 0.08 |
ENST00000331251.3 |
NPBWR1 |
neuropeptides B/W receptor 1 |
| chr7_-_80551671 | 0.08 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_-_8218997 | 0.08 |
ENST00000307637.4 |
C3AR1 |
complement component 3a receptor 1 |
| chr4_-_143226979 | 0.08 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chrX_-_30885319 | 0.08 |
ENST00000378933.1 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr8_-_124279627 | 0.08 |
ENST00000357082.4 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
| chr1_+_196743943 | 0.08 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chrX_+_153991025 | 0.08 |
ENST00000369550.5 |
DKC1 |
dyskeratosis congenita 1, dyskerin |
| chr8_+_21911054 | 0.08 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr4_-_155511887 | 0.08 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr7_+_16566449 | 0.08 |
ENST00000401542.2 |
LRRC72 |
leucine rich repeat containing 72 |
| chr16_-_4665023 | 0.08 |
ENST00000591897.1 |
UBALD1 |
UBA-like domain containing 1 |
| chr16_-_67597789 | 0.08 |
ENST00000605277.1 |
CTD-2012K14.6 |
CTD-2012K14.6 |
| chr15_+_69307028 | 0.07 |
ENST00000388866.3 ENST00000530406.2 |
NOX5 |
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr2_-_69180012 | 0.07 |
ENST00000481498.1 |
GKN2 |
gastrokine 2 |
| chr1_-_158301312 | 0.07 |
ENST00000368168.3 |
CD1B |
CD1b molecule |
| chr2_-_74007193 | 0.07 |
ENST00000377706.4 ENST00000443070.1 ENST00000272444.3 |
DUSP11 |
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr18_-_10787140 | 0.07 |
ENST00000383408.2 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr1_-_100643765 | 0.07 |
ENST00000370137.1 ENST00000370138.1 ENST00000342895.3 |
LRRC39 |
leucine rich repeat containing 39 |
| chr1_+_64014588 | 0.07 |
ENST00000371086.2 ENST00000340052.3 |
DLEU2L |
deleted in lymphocytic leukemia 2-like |
| chr7_-_24797546 | 0.07 |
ENST00000414428.1 ENST00000419307.1 ENST00000342947.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chrX_-_72097698 | 0.07 |
ENST00000373530.1 |
DMRTC1 |
DMRT-like family C1 |
| chr11_+_60197040 | 0.07 |
ENST00000300190.2 |
MS4A5 |
membrane-spanning 4-domains, subfamily A, member 5 |
| chr13_-_41837620 | 0.07 |
ENST00000379477.1 ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1 |
mitochondrial translational release factor 1 |
| chr14_+_39703112 | 0.07 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr10_-_75118471 | 0.07 |
ENST00000340329.3 |
TTC18 |
tetratricopeptide repeat domain 18 |
| chr10_-_62332357 | 0.07 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_+_101988627 | 0.07 |
ENST00000547405.1 ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr1_-_43424500 | 0.07 |
ENST00000415851.2 ENST00000426263.3 ENST00000372500.3 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr3_-_24207039 | 0.07 |
ENST00000280696.5 |
THRB |
thyroid hormone receptor, beta |
| chr16_-_4664860 | 0.07 |
ENST00000587615.1 ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1 |
UBA-like domain containing 1 |
| chr4_-_84035868 | 0.07 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr4_+_70796784 | 0.07 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr4_-_177116772 | 0.07 |
ENST00000280191.2 |
SPATA4 |
spermatogenesis associated 4 |
| chr3_-_87325612 | 0.07 |
ENST00000561167.1 ENST00000560656.1 ENST00000344265.3 |
POU1F1 |
POU class 1 homeobox 1 |
| chr4_+_187148556 | 0.07 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr12_+_48099858 | 0.06 |
ENST00000547799.1 |
RP1-197B17.3 |
RP1-197B17.3 |
| chr22_-_38699003 | 0.06 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chr4_-_153274078 | 0.06 |
ENST00000263981.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_88532028 | 0.06 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr1_-_197036364 | 0.06 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr4_-_84035905 | 0.06 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr7_+_107220422 | 0.06 |
ENST00000005259.4 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr11_+_73661364 | 0.06 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr22_-_45608324 | 0.06 |
ENST00000496226.1 ENST00000251993.7 |
KIAA0930 |
KIAA0930 |
| chr10_+_5005598 | 0.06 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr17_+_68071458 | 0.06 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_87325728 | 0.06 |
ENST00000350375.2 |
POU1F1 |
POU class 1 homeobox 1 |
| chr7_+_39125365 | 0.06 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr5_-_131879205 | 0.06 |
ENST00000231454.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr14_+_39583427 | 0.06 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr2_+_115219171 | 0.06 |
ENST00000409163.1 |
DPP10 |
dipeptidyl-peptidase 10 (non-functional) |
| chr6_+_26199737 | 0.06 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr1_+_117963209 | 0.06 |
ENST00000449370.2 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
| chr4_+_119199864 | 0.06 |
ENST00000602414.1 ENST00000602520.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr4_+_55095428 | 0.06 |
ENST00000508170.1 ENST00000512143.1 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr15_-_59981479 | 0.06 |
ENST00000607373.1 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr1_+_22962948 | 0.06 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr18_-_69246186 | 0.06 |
ENST00000568095.1 |
RP11-510D19.1 |
RP11-510D19.1 |
| chr17_+_53342311 | 0.06 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr6_+_30585486 | 0.06 |
ENST00000259873.4 ENST00000506373.2 |
MRPS18B |
mitochondrial ribosomal protein S18B |
| chr7_+_116593433 | 0.06 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr10_-_61720640 | 0.06 |
ENST00000521074.1 ENST00000444900.1 |
C10orf40 |
chromosome 10 open reading frame 40 |
| chr10_-_52645379 | 0.06 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr19_+_33865218 | 0.06 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr3_+_119217376 | 0.06 |
ENST00000494664.1 ENST00000493694.1 |
TIMMDC1 |
translocase of inner mitochondrial membrane domain containing 1 |
| chr20_+_15177480 | 0.06 |
ENST00000402914.1 |
MACROD2 |
MACRO domain containing 2 |
| chr10_-_126716459 | 0.06 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr11_+_69061594 | 0.06 |
ENST00000441339.2 ENST00000308946.3 ENST00000535407.1 |
MYEOV |
myeloma overexpressed |
| chr3_-_142297668 | 0.06 |
ENST00000350721.4 ENST00000383101.3 |
ATR |
ataxia telangiectasia and Rad3 related |
| chr18_-_19994830 | 0.06 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr3_+_130569592 | 0.06 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_76251879 | 0.06 |
ENST00000535300.1 ENST00000319942.3 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
| chr3_+_94657086 | 0.05 |
ENST00000463200.1 |
LINC00879 |
long intergenic non-protein coding RNA 879 |
| chr6_-_74161977 | 0.05 |
ENST00000370318.1 ENST00000370315.3 |
MB21D1 |
Mab-21 domain containing 1 |
| chr1_+_196946680 | 0.05 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr2_-_208989225 | 0.05 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr4_+_119199904 | 0.05 |
ENST00000602483.1 ENST00000602819.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr11_+_34664014 | 0.05 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr3_+_101280677 | 0.05 |
ENST00000309922.6 ENST00000495642.1 |
TRMT10C |
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr6_-_136571400 | 0.05 |
ENST00000418509.2 ENST00000420702.1 ENST00000451457.2 |
MTFR2 |
mitochondrial fission regulator 2 |
| chr7_+_112262421 | 0.05 |
ENST00000453459.1 |
AC002463.3 |
AC002463.3 |
| chr4_+_184826418 | 0.05 |
ENST00000308497.4 ENST00000438269.1 |
STOX2 |
storkhead box 2 |
| chr5_+_61874562 | 0.05 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr2_+_201754050 | 0.05 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr7_+_95115210 | 0.05 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr3_-_74570291 | 0.05 |
ENST00000263665.6 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
| chr2_+_201754135 | 0.05 |
ENST00000409357.1 ENST00000409129.2 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr5_-_86534822 | 0.05 |
ENST00000445770.2 |
AC008394.1 |
Uncharacterized protein |
| chr6_+_15401075 | 0.05 |
ENST00000541660.1 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr4_+_106631966 | 0.05 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr1_+_220267429 | 0.05 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr6_-_26056695 | 0.05 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr8_-_114389353 | 0.05 |
ENST00000343508.3 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr17_-_10452929 | 0.05 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.3 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 1.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016426 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |