Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for BACH2
Z-value: 0.16
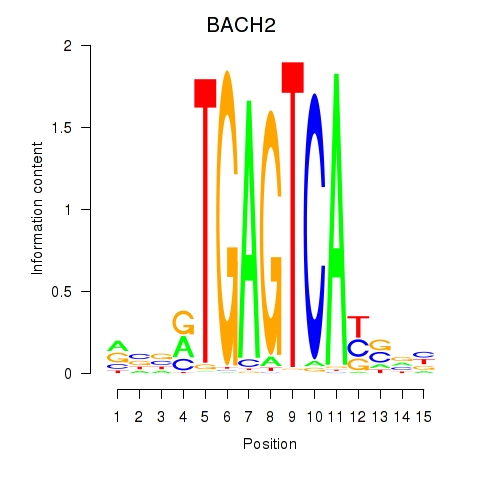
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BACH2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH2 | hg19_v2_chr6_-_91006461_91006517, hg19_v2_chr6_-_91006627_91006641 | -0.43 | 2.9e-01 | Click! |
Activity profile of BACH2 motif
Sorted Z-values of BACH2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_75919253 | 0.16 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr13_-_67802549 | 0.14 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr1_+_156096336 | 0.11 |
ENST00000504687.1 ENST00000473598.2 |
LMNA |
lamin A/C |
| chr6_-_24877490 | 0.11 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr12_+_7023735 | 0.10 |
ENST00000538763.1 ENST00000544774.1 ENST00000545045.2 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr12_+_7023491 | 0.10 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr2_-_219858123 | 0.10 |
ENST00000453769.1 ENST00000295728.2 ENST00000392096.2 |
CRYBA2 |
crystallin, beta A2 |
| chr16_+_15596123 | 0.09 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr11_-_66104237 | 0.09 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr17_-_76921459 | 0.09 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr5_+_110559784 | 0.09 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr16_+_53133070 | 0.08 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr20_+_34742650 | 0.08 |
ENST00000373945.1 ENST00000338074.2 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr11_-_65150103 | 0.08 |
ENST00000294187.6 ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr5_+_176237478 | 0.08 |
ENST00000329542.4 |
UNC5A |
unc-5 homolog A (C. elegans) |
| chr11_-_102826434 | 0.08 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr12_-_56101647 | 0.08 |
ENST00000347027.6 ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7 |
integrin, alpha 7 |
| chr19_-_46285646 | 0.07 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr11_-_66103932 | 0.07 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr17_-_33390667 | 0.07 |
ENST00000378516.2 ENST00000268850.7 ENST00000394597.2 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr6_-_56819385 | 0.07 |
ENST00000370754.5 ENST00000449297.2 |
DST |
dystonin |
| chr7_+_150065278 | 0.07 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr16_+_69599899 | 0.07 |
ENST00000567239.1 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr5_-_176923803 | 0.07 |
ENST00000506161.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr2_+_169926047 | 0.07 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr1_+_156084461 | 0.07 |
ENST00000347559.2 ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA |
lamin A/C |
| chr3_+_48507621 | 0.07 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr22_+_23077065 | 0.06 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr4_+_123747834 | 0.06 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr3_+_11178779 | 0.06 |
ENST00000438284.2 |
HRH1 |
histamine receptor H1 |
| chr5_-_176923846 | 0.06 |
ENST00000506537.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr3_+_48507210 | 0.06 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr8_+_104384616 | 0.06 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr16_+_58533951 | 0.06 |
ENST00000566192.1 ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4 |
NDRG family member 4 |
| chr2_-_89459813 | 0.06 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr5_-_75919217 | 0.06 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr3_-_187454281 | 0.06 |
ENST00000232014.4 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr5_-_141061759 | 0.06 |
ENST00000508305.1 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr19_-_51587502 | 0.06 |
ENST00000156499.2 ENST00000391802.1 |
KLK14 |
kallikrein-related peptidase 14 |
| chr20_+_48429356 | 0.06 |
ENST00000361573.2 ENST00000541138.1 ENST00000539601.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr20_+_48429233 | 0.06 |
ENST00000417961.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr5_-_141061777 | 0.06 |
ENST00000239440.4 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chrX_-_74743080 | 0.05 |
ENST00000373367.3 |
ZDHHC15 |
zinc finger, DHHC-type containing 15 |
| chr11_-_66103867 | 0.05 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr19_-_18995029 | 0.05 |
ENST00000596048.1 |
CERS1 |
ceramide synthase 1 |
| chr16_+_89988259 | 0.05 |
ENST00000554444.1 ENST00000556565.1 |
TUBB3 |
Tubulin beta-3 chain |
| chr17_-_7123021 | 0.05 |
ENST00000399510.2 |
DLG4 |
discs, large homolog 4 (Drosophila) |
| chr5_+_138089100 | 0.05 |
ENST00000520339.1 ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr5_+_147582348 | 0.05 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr5_+_110559603 | 0.05 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr17_+_4853442 | 0.05 |
ENST00000522301.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr22_+_22681656 | 0.05 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr5_+_159436120 | 0.05 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr9_-_35112376 | 0.05 |
ENST00000488109.2 |
FAM214B |
family with sequence similarity 214, member B |
| chr16_+_30077055 | 0.05 |
ENST00000564595.2 ENST00000569798.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr1_+_150480551 | 0.05 |
ENST00000369049.4 ENST00000369047.4 |
ECM1 |
extracellular matrix protein 1 |
| chr13_+_32838801 | 0.05 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr4_+_123747979 | 0.05 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr9_-_139922726 | 0.05 |
ENST00000265662.5 ENST00000371605.3 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr8_-_26724784 | 0.05 |
ENST00000380573.3 |
ADRA1A |
adrenoceptor alpha 1A |
| chr1_-_179112189 | 0.05 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_-_31161380 | 0.05 |
ENST00000569305.1 ENST00000418068.2 ENST00000268281.4 |
PRSS36 |
protease, serine, 36 |
| chr1_+_223889285 | 0.05 |
ENST00000433674.2 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr15_+_75335604 | 0.05 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr8_-_38386175 | 0.05 |
ENST00000437935.2 ENST00000358138.1 |
C8orf86 |
chromosome 8 open reading frame 86 |
| chr10_+_17270214 | 0.05 |
ENST00000544301.1 |
VIM |
vimentin |
| chr15_+_51633826 | 0.04 |
ENST00000335449.6 |
GLDN |
gliomedin |
| chr16_+_30077098 | 0.04 |
ENST00000395240.3 ENST00000566846.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr9_-_139922631 | 0.04 |
ENST00000341511.6 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr3_+_191046810 | 0.04 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr1_+_150480576 | 0.04 |
ENST00000346569.6 |
ECM1 |
extracellular matrix protein 1 |
| chr3_-_48632593 | 0.04 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr1_-_158656488 | 0.04 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr11_-_65149422 | 0.04 |
ENST00000526432.1 ENST00000527174.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr1_+_156095951 | 0.04 |
ENST00000448611.2 ENST00000368297.1 |
LMNA |
lamin A/C |
| chr22_+_22712087 | 0.04 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chr3_-_71834318 | 0.04 |
ENST00000353065.3 |
PROK2 |
prokineticin 2 |
| chr1_+_26606608 | 0.04 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr3_-_48601206 | 0.04 |
ENST00000273610.3 |
UCN2 |
urocortin 2 |
| chr5_+_179247759 | 0.04 |
ENST00000389805.4 ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1 |
sequestosome 1 |
| chr7_+_30960915 | 0.04 |
ENST00000441328.2 ENST00000409899.1 ENST00000409611.1 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chrX_+_12993202 | 0.04 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr3_-_56502375 | 0.04 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr9_-_130637244 | 0.04 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr11_-_6341724 | 0.04 |
ENST00000530979.1 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr16_-_1429627 | 0.04 |
ENST00000248104.7 |
UNKL |
unkempt family zinc finger-like |
| chr1_-_109968973 | 0.04 |
ENST00000271308.4 ENST00000538610.1 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr1_-_27816641 | 0.04 |
ENST00000430629.2 |
WASF2 |
WAS protein family, member 2 |
| chr9_+_4985016 | 0.04 |
ENST00000539801.1 |
JAK2 |
Janus kinase 2 |
| chr17_+_7123125 | 0.04 |
ENST00000356839.5 ENST00000583312.1 ENST00000350303.5 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
| chr9_+_4985228 | 0.04 |
ENST00000381652.3 |
JAK2 |
Janus kinase 2 |
| chr11_-_6341844 | 0.04 |
ENST00000303927.3 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr17_+_76311791 | 0.04 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr19_-_10450287 | 0.04 |
ENST00000589261.1 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr10_+_127661942 | 0.04 |
ENST00000417114.1 ENST00000445510.1 ENST00000368691.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr2_-_145275211 | 0.04 |
ENST00000462355.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr9_-_91793675 | 0.04 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr15_-_83316087 | 0.03 |
ENST00000568757.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_223853348 | 0.03 |
ENST00000366872.5 |
CAPN8 |
calpain 8 |
| chr5_-_138534071 | 0.03 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr1_-_11918988 | 0.03 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr16_+_69600058 | 0.03 |
ENST00000393742.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr1_+_26605618 | 0.03 |
ENST00000270792.5 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_+_47835404 | 0.03 |
ENST00000595464.1 |
C5AR2 |
complement component 5a receptor 2 |
| chr19_+_36630454 | 0.03 |
ENST00000246533.3 |
CAPNS1 |
calpain, small subunit 1 |
| chr19_+_36630961 | 0.03 |
ENST00000587718.1 ENST00000592483.1 ENST00000590874.1 ENST00000588815.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr12_-_91348949 | 0.03 |
ENST00000358859.2 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
| chr7_-_150721570 | 0.03 |
ENST00000377974.2 ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B |
autophagy related 9B |
| chr11_+_57308979 | 0.03 |
ENST00000457912.1 |
SMTNL1 |
smoothelin-like 1 |
| chr3_-_98241713 | 0.03 |
ENST00000502288.1 ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1 |
claudin domain containing 1 |
| chr16_+_69600209 | 0.03 |
ENST00000566899.1 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr12_-_56122426 | 0.03 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr1_-_151965048 | 0.03 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr15_-_99789736 | 0.03 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr19_+_36630855 | 0.03 |
ENST00000589146.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr10_+_88428370 | 0.03 |
ENST00000372066.3 ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3 |
LIM domain binding 3 |
| chr7_+_134528635 | 0.03 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr16_-_1429674 | 0.03 |
ENST00000403703.1 ENST00000397464.1 ENST00000402641.2 |
UNKL |
unkempt family zinc finger-like |
| chr6_+_74405804 | 0.03 |
ENST00000287097.5 |
CD109 |
CD109 molecule |
| chr10_+_88854926 | 0.03 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr17_+_30677136 | 0.03 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr5_-_141030943 | 0.03 |
ENST00000522783.1 ENST00000519800.1 ENST00000435817.2 |
FCHSD1 |
FCH and double SH3 domains 1 |
| chr17_-_47287928 | 0.03 |
ENST00000507680.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr6_+_56820018 | 0.03 |
ENST00000370746.3 |
BEND6 |
BEN domain containing 6 |
| chr2_+_87754887 | 0.03 |
ENST00000409054.1 ENST00000331944.6 ENST00000409139.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr8_+_22844913 | 0.03 |
ENST00000519685.1 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr2_+_87754989 | 0.03 |
ENST00000409898.2 ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr6_+_292051 | 0.03 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr6_+_74405501 | 0.03 |
ENST00000437994.2 ENST00000422508.2 |
CD109 |
CD109 molecule |
| chr2_+_87755054 | 0.03 |
ENST00000423846.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr15_-_60690163 | 0.03 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr12_-_76478686 | 0.03 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr12_-_6809958 | 0.03 |
ENST00000320591.5 ENST00000534837.1 |
PIANP |
PILR alpha associated neural protein |
| chr6_+_56819773 | 0.03 |
ENST00000370750.2 |
BEND6 |
BEN domain containing 6 |
| chr17_-_38911580 | 0.03 |
ENST00000312150.4 |
KRT25 |
keratin 25 |
| chr9_-_130341268 | 0.03 |
ENST00000373314.3 |
FAM129B |
family with sequence similarity 129, member B |
| chr1_-_179112173 | 0.03 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr9_-_114246635 | 0.03 |
ENST00000338205.5 |
KIAA0368 |
KIAA0368 |
| chr11_-_47207390 | 0.03 |
ENST00000539589.1 ENST00000528462.1 |
PACSIN3 |
protein kinase C and casein kinase substrate in neurons 3 |
| chr12_-_39299406 | 0.03 |
ENST00000331366.5 |
CPNE8 |
copine VIII |
| chr3_+_187930719 | 0.03 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr16_-_89043377 | 0.02 |
ENST00000436887.2 ENST00000448839.1 ENST00000360302.2 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr1_+_156863470 | 0.02 |
ENST00000338302.3 ENST00000455314.1 ENST00000292357.7 |
PEAR1 |
platelet endothelial aggregation receptor 1 |
| chr7_-_98467629 | 0.02 |
ENST00000339375.4 |
TMEM130 |
transmembrane protein 130 |
| chr7_-_98467543 | 0.02 |
ENST00000345589.4 |
TMEM130 |
transmembrane protein 130 |
| chr11_+_46316677 | 0.02 |
ENST00000534787.1 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr6_+_2988847 | 0.02 |
ENST00000380472.3 ENST00000605901.1 ENST00000454015.1 |
NQO2 LINC01011 |
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr11_+_64692143 | 0.02 |
ENST00000164133.2 ENST00000532850.1 |
PPP2R5B |
protein phosphatase 2, regulatory subunit B', beta |
| chr17_+_30469579 | 0.02 |
ENST00000354266.3 ENST00000581094.1 ENST00000394692.2 |
RHOT1 |
ras homolog family member T1 |
| chr17_-_42345487 | 0.02 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr12_-_48152611 | 0.02 |
ENST00000389212.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr15_+_22892663 | 0.02 |
ENST00000313077.7 ENST00000561274.1 ENST00000560848.1 |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
| chr1_+_901847 | 0.02 |
ENST00000379410.3 ENST00000379409.2 ENST00000379407.3 |
PLEKHN1 |
pleckstrin homology domain containing, family N member 1 |
| chr10_-_31288398 | 0.02 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr17_-_73150629 | 0.02 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
| chr16_+_28996416 | 0.02 |
ENST00000395456.2 ENST00000454369.2 |
LAT |
linker for activation of T cells |
| chr2_+_201980827 | 0.02 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr20_-_634000 | 0.02 |
ENST00000381962.3 |
SRXN1 |
sulfiredoxin 1 |
| chr17_-_56595196 | 0.02 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr1_+_154966058 | 0.02 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr2_+_220143989 | 0.02 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr22_+_22735135 | 0.02 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr7_-_98467489 | 0.02 |
ENST00000416379.2 |
TMEM130 |
transmembrane protein 130 |
| chr12_-_57871853 | 0.02 |
ENST00000549602.1 ENST00000430041.2 |
ARHGAP9 |
Rho GTPase activating protein 9 |
| chr15_+_59903975 | 0.02 |
ENST00000560585.1 ENST00000396065.1 |
GCNT3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr6_-_31704282 | 0.02 |
ENST00000375784.3 ENST00000375779.2 |
CLIC1 |
chloride intracellular channel 1 |
| chr15_-_44829057 | 0.02 |
ENST00000559356.1 ENST00000560049.1 ENST00000313807.4 |
EIF3J-AS1 |
EIF3J antisense RNA 1 (head to head) |
| chr5_+_72143988 | 0.02 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr17_+_7590734 | 0.02 |
ENST00000457584.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr4_+_159727272 | 0.02 |
ENST00000379346.3 |
FNIP2 |
folliculin interacting protein 2 |
| chr5_-_137090028 | 0.02 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr14_-_23426322 | 0.02 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426337 | 0.02 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chrX_-_48901012 | 0.02 |
ENST00000315869.7 |
TFE3 |
transcription factor binding to IGHM enhancer 3 |
| chr2_+_220144052 | 0.02 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr22_+_22749343 | 0.02 |
ENST00000390298.2 |
IGLV7-43 |
immunoglobulin lambda variable 7-43 |
| chr14_-_23426270 | 0.02 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr3_-_81792780 | 0.02 |
ENST00000489715.1 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr12_-_48152853 | 0.02 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr2_+_231921574 | 0.02 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr2_-_65593784 | 0.02 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr22_+_22764088 | 0.02 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr1_-_173991434 | 0.02 |
ENST00000367696.2 |
RC3H1 |
ring finger and CCCH-type domains 1 |
| chr17_-_27045427 | 0.02 |
ENST00000301043.6 ENST00000412625.1 |
RAB34 |
RAB34, member RAS oncogene family |
| chrY_+_15016725 | 0.02 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr9_-_32573130 | 0.02 |
ENST00000350021.2 ENST00000379847.3 |
NDUFB6 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr7_+_99647389 | 0.02 |
ENST00000543588.1 ENST00000292450.4 ENST00000456748.2 |
ZSCAN21 |
zinc finger and SCAN domain containing 21 |
| chr17_+_79650962 | 0.02 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr12_-_719573 | 0.02 |
ENST00000397265.3 |
NINJ2 |
ninjurin 2 |
| chr17_+_66539369 | 0.02 |
ENST00000600820.1 |
AC079210.1 |
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chrX_+_41193407 | 0.02 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr4_-_74088800 | 0.02 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr3_-_49395705 | 0.02 |
ENST00000419349.1 |
GPX1 |
glutathione peroxidase 1 |
| chr3_-_194090460 | 0.02 |
ENST00000428839.1 ENST00000347624.3 |
LRRC15 |
leucine rich repeat containing 15 |
| chr2_-_85641162 | 0.02 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr12_+_6644443 | 0.02 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_223889310 | 0.02 |
ENST00000434648.1 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr17_-_27045405 | 0.02 |
ENST00000430132.2 |
RAB34 |
RAB34, member RAS oncogene family |
| chr7_-_96339132 | 0.02 |
ENST00000413065.1 |
SHFM1 |
split hand/foot malformation (ectrodactyly) type 1 |
| chr4_-_987217 | 0.02 |
ENST00000361661.2 ENST00000398516.2 |
SLC26A1 |
solute carrier family 26 (anion exchanger), member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.0 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.0 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.0 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.0 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |