Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for BATF3
Z-value: 0.11
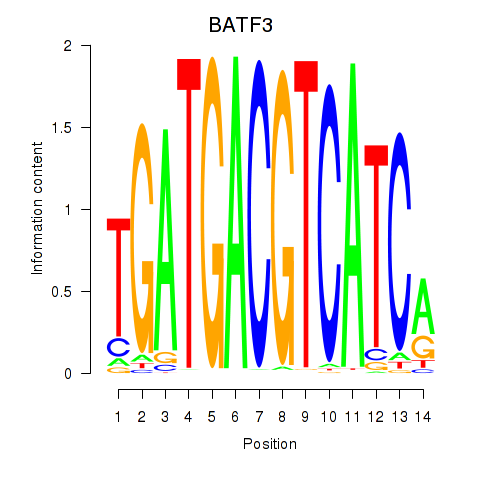
Transcription factors associated with BATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF3
|
ENSG00000123685.4 | BATF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BATF3 | hg19_v2_chr1_-_212873267_212873332 | -0.45 | 2.6e-01 | Click! |
Activity profile of BATF3 motif
Sorted Z-values of BATF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BATF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_37594185 | 0.13 |
ENST00000518586.1 ENST00000335171.6 ENST00000521644.1 |
ERLIN2 |
ER lipid raft associated 2 |
| chr8_+_37594103 | 0.12 |
ENST00000397228.2 |
ERLIN2 |
ER lipid raft associated 2 |
| chr4_-_104119528 | 0.12 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chr2_-_74757066 | 0.11 |
ENST00000377526.3 |
AUP1 |
ancient ubiquitous protein 1 |
| chr1_+_156030937 | 0.10 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chrX_-_135056106 | 0.10 |
ENST00000433339.2 |
MMGT1 |
membrane magnesium transporter 1 |
| chr15_+_34394257 | 0.09 |
ENST00000397766.2 |
PGBD4 |
piggyBac transposable element derived 4 |
| chr2_-_202645612 | 0.09 |
ENST00000409632.2 ENST00000410052.1 ENST00000467448.1 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
| chr18_+_76829441 | 0.09 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chr7_-_121944491 | 0.09 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr2_+_70142189 | 0.08 |
ENST00000264444.2 |
MXD1 |
MAX dimerization protein 1 |
| chr1_+_228353495 | 0.08 |
ENST00000366711.3 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr19_-_47616992 | 0.08 |
ENST00000253048.5 |
ZC3H4 |
zinc finger CCCH-type containing 4 |
| chr15_-_34394119 | 0.08 |
ENST00000256545.4 |
EMC7 |
ER membrane protein complex subunit 7 |
| chr15_-_34394008 | 0.07 |
ENST00000527822.1 ENST00000528949.1 |
EMC7 |
ER membrane protein complex subunit 7 |
| chr11_-_28129656 | 0.07 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr22_+_31518938 | 0.06 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr17_-_49124230 | 0.06 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr8_+_37594130 | 0.06 |
ENST00000518526.1 ENST00000523887.1 ENST00000276461.5 |
ERLIN2 |
ER lipid raft associated 2 |
| chr19_-_49622348 | 0.06 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chrX_-_135056216 | 0.06 |
ENST00000305963.2 |
MMGT1 |
membrane magnesium transporter 1 |
| chr12_+_7282795 | 0.05 |
ENST00000266546.6 |
CLSTN3 |
calsyntenin 3 |
| chr2_-_43823093 | 0.05 |
ENST00000405006.4 |
THADA |
thyroid adenoma associated |
| chr16_+_3068393 | 0.05 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr2_+_70142232 | 0.05 |
ENST00000540449.1 |
MXD1 |
MAX dimerization protein 1 |
| chr18_+_76829385 | 0.05 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chr19_+_18723660 | 0.05 |
ENST00000262817.3 |
TMEM59L |
transmembrane protein 59-like |
| chr22_+_38597889 | 0.05 |
ENST00000338483.2 ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chrX_+_83116142 | 0.05 |
ENST00000329312.4 |
CYLC1 |
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr2_-_43823119 | 0.05 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr14_+_68086515 | 0.05 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr1_+_151682909 | 0.04 |
ENST00000326413.3 ENST00000442233.2 |
RIIAD1 AL589765.1 |
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr16_+_68056844 | 0.04 |
ENST00000565263.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr3_-_119182523 | 0.04 |
ENST00000319172.5 |
TMEM39A |
transmembrane protein 39A |
| chr14_-_24732403 | 0.04 |
ENST00000206765.6 |
TGM1 |
transglutaminase 1 |
| chr16_+_68057179 | 0.04 |
ENST00000567100.1 ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr16_+_68057153 | 0.04 |
ENST00000358896.6 ENST00000568099.2 |
DUS2 |
dihydrouridine synthase 2 |
| chr11_+_66059339 | 0.04 |
ENST00000327259.4 |
TMEM151A |
transmembrane protein 151A |
| chr16_+_56225248 | 0.04 |
ENST00000262493.6 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr3_+_38206975 | 0.03 |
ENST00000446845.1 ENST00000311806.3 |
OXSR1 |
oxidative stress responsive 1 |
| chr5_+_112849373 | 0.03 |
ENST00000161863.4 ENST00000515883.1 |
YTHDC2 |
YTH domain containing 2 |
| chr10_+_123923105 | 0.03 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr10_+_123922941 | 0.03 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr10_+_123923205 | 0.03 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr17_+_7835419 | 0.03 |
ENST00000576538.1 ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB |
centrobin, centrosomal BRCA2 interacting protein |
| chr6_+_37400974 | 0.03 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr6_+_32939964 | 0.03 |
ENST00000607833.1 |
BRD2 |
bromodomain containing 2 |
| chr2_-_202645835 | 0.02 |
ENST00000264276.6 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
| chr11_-_85376121 | 0.02 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chr11_+_102217936 | 0.02 |
ENST00000532832.1 ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2 |
baculoviral IAP repeat containing 2 |
| chr1_-_70671216 | 0.02 |
ENST00000370952.3 |
LRRC40 |
leucine rich repeat containing 40 |
| chr19_-_460996 | 0.02 |
ENST00000264554.6 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr9_-_100684845 | 0.02 |
ENST00000375119.3 |
C9orf156 |
chromosome 9 open reading frame 156 |
| chr17_+_44076616 | 0.02 |
ENST00000537309.1 |
STH |
saitohin |
| chr10_-_128210005 | 0.01 |
ENST00000284694.7 ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr17_+_29421987 | 0.01 |
ENST00000431387.4 |
NF1 |
neurofibromin 1 |
| chr8_-_67090825 | 0.01 |
ENST00000276571.3 |
CRH |
corticotropin releasing hormone |
| chr9_-_99381660 | 0.01 |
ENST00000375240.3 ENST00000463569.1 |
CDC14B |
cell division cycle 14B |
| chr13_-_36871886 | 0.01 |
ENST00000491049.2 ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169 SOHLH2 CCDC169-SOHLH2 |
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr14_-_24732368 | 0.01 |
ENST00000544573.1 |
TGM1 |
transglutaminase 1 |
| chr11_-_65793948 | 0.01 |
ENST00000312106.5 |
CATSPER1 |
cation channel, sperm associated 1 |
| chr4_-_2010562 | 0.01 |
ENST00000411649.1 ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA |
negative elongation factor complex member A |
| chr3_+_128598433 | 0.00 |
ENST00000308982.7 ENST00000514336.1 |
ACAD9 |
acyl-CoA dehydrogenase family, member 9 |
| chr1_+_153750622 | 0.00 |
ENST00000532853.1 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr2_+_74757050 | 0.00 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr15_+_42841008 | 0.00 |
ENST00000260372.3 ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2 |
HAUS augmin-like complex, subunit 2 |
| chr1_+_70671363 | 0.00 |
ENST00000370951.1 ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11 |
serine/arginine-rich splicing factor 11 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.0 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |