Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 1.55
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
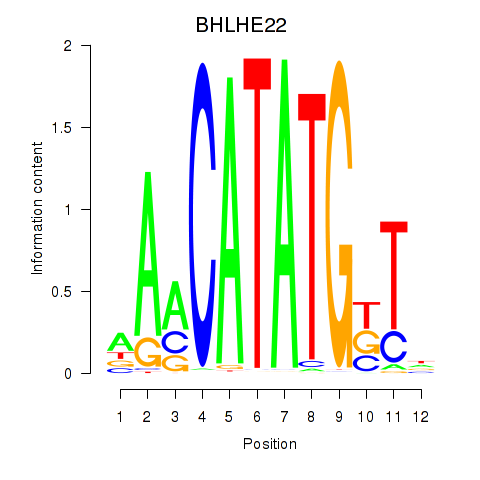
BHLHE22
|
ENSG00000180828.1 | BHLHE22 |
|
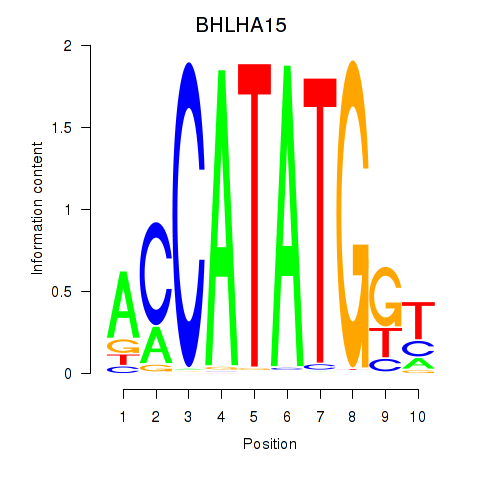
BHLHA15
|
ENSG00000180535.3 | BHLHA15 |
|
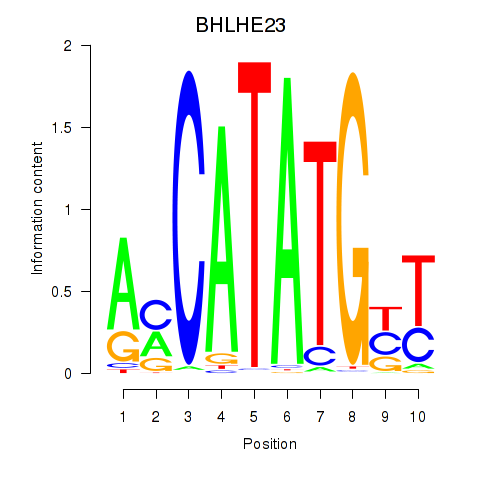
BHLHE23
|
ENSG00000125533.4 | BHLHE23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE23 | hg19_v2_chr20_-_61638313_61638387 | -0.72 | 4.5e-02 | Click! |
| BHLHE22 | hg19_v2_chr8_+_65492756_65492814 | -0.50 | 2.1e-01 | Click! |
Activity profile of BHLHE22_BHLHA15_BHLHE23 motif
Sorted Z-values of BHLHE22_BHLHA15_BHLHE23 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE22_BHLHA15_BHLHE23
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111312622 | 3.52 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr2_-_188378368 | 2.31 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr9_+_18474098 | 1.25 |
ENST00000327883.7 ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr12_-_91573132 | 1.03 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr1_+_170633047 | 1.01 |
ENST00000239461.6 ENST00000497230.2 |
PRRX1 |
paired related homeobox 1 |
| chr12_-_91573316 | 0.98 |
ENST00000393155.1 |
DCN |
decorin |
| chr3_-_114477962 | 0.92 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_-_114477787 | 0.90 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr13_+_49551020 | 0.81 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr4_+_160188889 | 0.80 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_163099885 | 0.69 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr4_+_55095264 | 0.51 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chrX_+_70798261 | 0.51 |
ENST00000373696.3 |
ACRC |
acidic repeat containing |
| chr1_-_186344802 | 0.51 |
ENST00000451586.1 |
TPR |
translocated promoter region, nuclear basket protein |
| chr15_-_98417780 | 0.47 |
ENST00000503874.3 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
| chr12_-_108714412 | 0.47 |
ENST00000412676.1 ENST00000550573.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr2_+_210444748 | 0.46 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr10_-_31288398 | 0.46 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr1_+_104293028 | 0.44 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr19_+_18208603 | 0.41 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr6_+_69942298 | 0.41 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr3_-_178984759 | 0.40 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_-_55700216 | 0.38 |
ENST00000569205.1 |
CCPG1 |
cell cycle progression 1 |
| chr4_-_159094194 | 0.38 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr3_-_18480260 | 0.35 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr15_-_55700522 | 0.34 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr7_-_38403077 | 0.33 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr5_-_137674000 | 0.33 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr6_+_73076432 | 0.32 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr2_-_163100045 | 0.32 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr2_-_182545603 | 0.31 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr3_-_121379739 | 0.31 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr1_+_61330931 | 0.30 |
ENST00000371191.1 |
NFIA |
nuclear factor I/A |
| chr1_+_95975672 | 0.29 |
ENST00000440116.2 ENST00000456933.1 |
RP11-286B14.1 |
RP11-286B14.1 |
| chr19_-_37019562 | 0.29 |
ENST00000523638.1 |
ZNF260 |
zinc finger protein 260 |
| chr16_-_4896205 | 0.29 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr15_-_63448973 | 0.28 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr7_+_148287657 | 0.26 |
ENST00000307003.2 |
C7orf33 |
chromosome 7 open reading frame 33 |
| chr5_+_173316341 | 0.25 |
ENST00000520867.1 ENST00000334035.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr16_+_21716284 | 0.25 |
ENST00000388957.3 |
OTOA |
otoancorin |
| chr9_+_18474163 | 0.25 |
ENST00000380566.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr12_-_91573249 | 0.23 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr5_+_135496675 | 0.23 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr3_+_185300391 | 0.22 |
ENST00000545472.1 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr15_-_55700457 | 0.19 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr6_-_169364429 | 0.19 |
ENST00000444586.1 |
RP3-495K2.3 |
RP3-495K2.3 |
| chr10_+_5135981 | 0.19 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr5_+_157158205 | 0.18 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr7_-_97881429 | 0.17 |
ENST00000420697.1 ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
| chr6_-_112115074 | 0.17 |
ENST00000368667.2 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr11_+_65405556 | 0.16 |
ENST00000534313.1 ENST00000533361.1 ENST00000526137.1 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr15_+_26360970 | 0.15 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr2_-_183387430 | 0.14 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr9_-_130635741 | 0.14 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chrX_+_120181457 | 0.13 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr11_-_26743546 | 0.13 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_+_58166370 | 0.12 |
ENST00000300209.8 |
METTL21B |
methyltransferase like 21B |
| chr2_+_223162866 | 0.11 |
ENST00000295226.1 |
CCDC140 |
coiled-coil domain containing 140 |
| chr2_-_32490859 | 0.11 |
ENST00000404025.2 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr8_-_23315190 | 0.10 |
ENST00000356206.6 ENST00000358689.4 ENST00000417069.2 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_-_171433579 | 0.10 |
ENST00000265094.5 ENST00000393802.2 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr8_-_135522425 | 0.09 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr2_-_32490801 | 0.09 |
ENST00000360906.5 ENST00000342905.6 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr9_+_2158443 | 0.09 |
ENST00000302401.3 ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_157670528 | 0.09 |
ENST00000368186.5 ENST00000496769.1 |
FCRL3 |
Fc receptor-like 3 |
| chr9_+_2158485 | 0.09 |
ENST00000417599.1 ENST00000382185.1 ENST00000382183.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_+_60758814 | 0.09 |
ENST00000579432.1 ENST00000446119.2 |
MRC2 |
mannose receptor, C type 2 |
| chr20_-_34117447 | 0.08 |
ENST00000246199.2 ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173 |
chromosome 20 open reading frame 173 |
| chr12_+_58166431 | 0.08 |
ENST00000333012.5 |
METTL21B |
methyltransferase like 21B |
| chr9_+_12775011 | 0.08 |
ENST00000319264.3 |
LURAP1L |
leucine rich adaptor protein 1-like |
| chr4_+_114214125 | 0.08 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chrX_-_13835147 | 0.08 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr1_+_100810575 | 0.08 |
ENST00000542213.1 |
CDC14A |
cell division cycle 14A |
| chr5_-_171433819 | 0.08 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr3_+_62304648 | 0.08 |
ENST00000462069.1 ENST00000232519.5 ENST00000465142.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr10_+_133918175 | 0.08 |
ENST00000298622.4 |
JAKMIP3 |
Janus kinase and microtubule interacting protein 3 |
| chr20_-_30060816 | 0.08 |
ENST00000317676.2 |
DEFB124 |
defensin, beta 124 |
| chr16_+_31119615 | 0.08 |
ENST00000394950.3 ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK |
branched chain ketoacid dehydrogenase kinase |
| chr10_-_48416849 | 0.07 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr4_-_74088800 | 0.07 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr14_+_77292715 | 0.07 |
ENST00000393774.3 ENST00000555189.1 ENST00000450042.2 |
C14orf166B |
chromosome 14 open reading frame 166B |
| chr2_-_183387283 | 0.07 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_97010424 | 0.07 |
ENST00000541107.1 ENST00000450218.1 ENST00000326771.2 |
FHL5 |
four and a half LIM domains 5 |
| chr3_+_137717571 | 0.07 |
ENST00000343735.4 |
CLDN18 |
claudin 18 |
| chr1_+_159272111 | 0.06 |
ENST00000368114.1 |
FCER1A |
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr1_+_12524965 | 0.06 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr19_+_56368803 | 0.06 |
ENST00000587891.1 |
NLRP4 |
NLR family, pyrin domain containing 4 |
| chr6_-_119031228 | 0.06 |
ENST00000392500.3 ENST00000368488.5 ENST00000434604.1 |
CEP85L |
centrosomal protein 85kDa-like |
| chr3_+_171758344 | 0.06 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chrX_-_13835461 | 0.05 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr4_+_25915896 | 0.05 |
ENST00000514384.1 |
SMIM20 |
small integral membrane protein 20 |
| chr2_+_186603355 | 0.05 |
ENST00000343098.5 |
FSIP2 |
fibrous sheath interacting protein 2 |
| chr9_+_108424738 | 0.05 |
ENST00000334077.3 |
TAL2 |
T-cell acute lymphocytic leukemia 2 |
| chr15_-_65503801 | 0.05 |
ENST00000261883.4 |
CILP |
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr7_-_99573677 | 0.05 |
ENST00000292401.4 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr4_+_2814011 | 0.05 |
ENST00000502260.1 ENST00000435136.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr11_-_88796803 | 0.05 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chr3_+_137728842 | 0.05 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr1_+_50569575 | 0.05 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr12_-_95397442 | 0.04 |
ENST00000547157.1 ENST00000547986.1 ENST00000327772.2 |
NDUFA12 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chrX_+_107334895 | 0.04 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr5_-_114632307 | 0.04 |
ENST00000506442.1 ENST00000379611.5 |
CCDC112 |
coiled-coil domain containing 112 |
| chr15_+_63354769 | 0.04 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr12_+_50794730 | 0.04 |
ENST00000523389.1 ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4 |
La ribonucleoprotein domain family, member 4 |
| chr2_-_183387064 | 0.04 |
ENST00000536095.1 ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_116502605 | 0.04 |
ENST00000458284.2 ENST00000490693.1 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr2_+_108994633 | 0.04 |
ENST00000409309.3 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_+_47489240 | 0.04 |
ENST00000371901.3 |
CYP4X1 |
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr11_+_55650773 | 0.04 |
ENST00000449290.2 |
TRIM51 |
tripartite motif-containing 51 |
| chr2_+_108994466 | 0.03 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr8_-_67976509 | 0.03 |
ENST00000518747.1 |
COPS5 |
COP9 signalosome subunit 5 |
| chr20_+_57226841 | 0.03 |
ENST00000358029.4 ENST00000361830.3 |
STX16 |
syntaxin 16 |
| chr4_+_2813946 | 0.03 |
ENST00000442312.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr21_-_36421401 | 0.03 |
ENST00000486278.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr14_-_69864993 | 0.02 |
ENST00000555373.1 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
| chr14_-_51297197 | 0.02 |
ENST00000382043.4 |
NIN |
ninein (GSK3B interacting protein) |
| chr11_+_120107344 | 0.02 |
ENST00000260264.4 |
POU2F3 |
POU class 2 homeobox 3 |
| chr10_+_51565108 | 0.02 |
ENST00000438493.1 ENST00000452682.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr19_+_57874835 | 0.02 |
ENST00000543226.1 ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1 ZNF547 AC003002.4 |
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr8_+_67976593 | 0.02 |
ENST00000262210.5 ENST00000412460.1 |
CSPP1 |
centrosome and spindle pole associated protein 1 |
| chr3_-_131753830 | 0.02 |
ENST00000429747.1 |
CPNE4 |
copine IV |
| chr7_+_116502527 | 0.02 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr3_-_46069223 | 0.02 |
ENST00000309285.3 |
XCR1 |
chemokine (C motif) receptor 1 |
| chr12_-_75723805 | 0.01 |
ENST00000409799.1 ENST00000409445.3 |
CAPS2 |
calcyphosine 2 |
| chr10_+_18629628 | 0.01 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr15_+_54901540 | 0.01 |
ENST00000539562.2 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr10_+_51565188 | 0.01 |
ENST00000430396.2 ENST00000374087.4 ENST00000414907.2 |
NCOA4 |
nuclear receptor coactivator 4 |
| chrX_+_69509927 | 0.01 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr6_+_37400974 | 0.01 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr1_-_99470558 | 0.01 |
ENST00000370188.3 |
LPPR5 |
Lipid phosphate phosphatase-related protein type 5 |
| chr2_+_181845843 | 0.00 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr5_+_156607829 | 0.00 |
ENST00000422843.3 |
ITK |
IL2-inducible T-cell kinase |
| chr7_+_107531580 | 0.00 |
ENST00000537148.1 ENST00000440410.1 ENST00000437604.2 |
DLD |
dihydrolipoamide dehydrogenase |
| chr1_-_44818599 | 0.00 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr4_-_113207048 | 0.00 |
ENST00000361717.3 |
TIFA |
TRAF-interacting protein with forkhead-associated domain |
| chr10_-_69597810 | 0.00 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_205091115 | 0.00 |
ENST00000264515.6 ENST00000367164.1 |
RBBP5 |
retinoblastoma binding protein 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 0.8 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 0.9 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.5 | GO:0045799 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 2.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.5 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |