Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for CDC5L
Z-value: 0.82
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | CDC5L |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | -0.62 | 9.9e-02 | Click! |
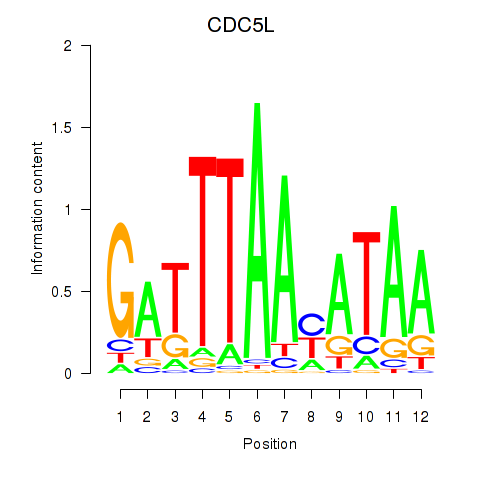
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22458631 | 1.02 |
ENST00000390444.1 |
TRAV16 |
T cell receptor alpha variable 16 |
| chr18_+_61554932 | 0.87 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr18_+_616672 | 0.86 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr1_+_62439037 | 0.81 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr18_+_616711 | 0.78 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr6_-_136847099 | 0.75 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr6_-_136847610 | 0.74 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr2_-_113542063 | 0.74 |
ENST00000263339.3 |
IL1A |
interleukin 1, alpha |
| chr19_-_10697895 | 0.66 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr17_-_7493390 | 0.64 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr18_+_61445007 | 0.62 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr18_+_61442629 | 0.62 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr5_+_167181917 | 0.61 |
ENST00000519204.1 |
TENM2 |
teneurin transmembrane protein 2 |
| chr3_+_122044084 | 0.58 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr11_+_69924639 | 0.56 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr8_-_57233103 | 0.53 |
ENST00000303749.3 ENST00000522671.1 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr1_+_152956549 | 0.52 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr5_+_140227048 | 0.51 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr4_-_110723194 | 0.46 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr6_-_155776966 | 0.45 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr4_-_110723134 | 0.44 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr8_+_99956759 | 0.44 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr5_+_140213815 | 0.43 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr10_-_116444371 | 0.42 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr18_-_28682374 | 0.42 |
ENST00000280904.6 |
DSC2 |
desmocollin 2 |
| chr9_+_35673853 | 0.41 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr8_-_124749609 | 0.41 |
ENST00000262219.6 ENST00000419625.1 |
ANXA13 |
annexin A13 |
| chr21_+_43619796 | 0.41 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr4_+_155484155 | 0.40 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr2_+_207804278 | 0.40 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr14_+_39703112 | 0.38 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr1_+_244515930 | 0.38 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr5_+_54398463 | 0.37 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr15_+_26360970 | 0.37 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr14_-_99737565 | 0.37 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_+_144339738 | 0.37 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr1_+_160370344 | 0.36 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr18_-_19994830 | 0.35 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr5_+_140220769 | 0.35 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr21_-_31852663 | 0.34 |
ENST00000390689.2 |
KRTAP19-1 |
keratin associated protein 19-1 |
| chr8_+_24772455 | 0.34 |
ENST00000433454.2 |
NEFM |
neurofilament, medium polypeptide |
| chr10_-_98031265 | 0.34 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr5_-_82969405 | 0.33 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr6_+_27791862 | 0.32 |
ENST00000355057.1 |
HIST1H4J |
histone cluster 1, H4j |
| chrX_-_24690771 | 0.32 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr2_+_18059906 | 0.32 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr6_-_53409890 | 0.32 |
ENST00000229416.6 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
| chr3_-_108672609 | 0.31 |
ENST00000393963.3 ENST00000471108.1 |
GUCA1C |
guanylate cyclase activator 1C |
| chr5_+_145316120 | 0.30 |
ENST00000359120.4 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr10_-_98031310 | 0.30 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr12_+_20968608 | 0.30 |
ENST00000381541.3 ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3 SLCO1B3 SLCO1B7 |
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_-_61495760 | 0.30 |
ENST00000395347.1 |
SLC16A9 |
solute carrier family 16, member 9 |
| chr5_+_140165876 | 0.29 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr12_+_21168630 | 0.29 |
ENST00000421593.2 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr14_-_99737822 | 0.28 |
ENST00000345514.2 ENST00000443726.2 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr8_-_86253888 | 0.28 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr8_+_104892639 | 0.28 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr12_-_26278030 | 0.28 |
ENST00000242728.4 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
| chr7_-_20256965 | 0.27 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr4_-_122302163 | 0.27 |
ENST00000394427.2 |
QRFPR |
pyroglutamylated RFamide peptide receptor |
| chr10_+_96698406 | 0.27 |
ENST00000260682.6 |
CYP2C9 |
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr8_-_15095832 | 0.27 |
ENST00000382080.1 |
SGCZ |
sarcoglycan, zeta |
| chr12_+_21207503 | 0.27 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_+_57881966 | 0.26 |
ENST00000495364.1 |
SLMAP |
sarcolemma associated protein |
| chr6_-_39197226 | 0.26 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr2_+_103353367 | 0.26 |
ENST00000454536.1 ENST00000409528.1 ENST00000409173.1 |
TMEM182 |
transmembrane protein 182 |
| chr10_-_116286563 | 0.25 |
ENST00000369253.2 |
ABLIM1 |
actin binding LIM protein 1 |
| chr12_-_39734783 | 0.25 |
ENST00000552961.1 |
KIF21A |
kinesin family member 21A |
| chr8_+_21911054 | 0.25 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr7_-_16921601 | 0.25 |
ENST00000402239.3 ENST00000310398.2 ENST00000414935.1 |
AGR3 |
anterior gradient 3 |
| chr2_+_238877424 | 0.25 |
ENST00000434655.1 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
| chr3_+_35721182 | 0.25 |
ENST00000413378.1 ENST00000417925.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr5_+_140180635 | 0.25 |
ENST00000522353.2 ENST00000532566.2 |
PCDHA3 |
protocadherin alpha 3 |
| chr16_+_4845379 | 0.24 |
ENST00000588606.1 ENST00000586005.1 |
SMIM22 |
small integral membrane protein 22 |
| chr6_-_32557610 | 0.24 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr8_+_98900132 | 0.23 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr6_+_121756809 | 0.23 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr1_-_226926864 | 0.23 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr15_-_22448819 | 0.23 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr13_-_86373536 | 0.23 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr14_+_70918874 | 0.23 |
ENST00000603540.1 |
ADAM21 |
ADAM metallopeptidase domain 21 |
| chr8_+_75736761 | 0.23 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr3_-_119396193 | 0.23 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr9_-_19786926 | 0.23 |
ENST00000341998.2 ENST00000286344.3 |
SLC24A2 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr12_-_87232644 | 0.23 |
ENST00000549405.2 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr9_+_135854091 | 0.23 |
ENST00000450530.1 ENST00000534944.1 |
GFI1B |
growth factor independent 1B transcription repressor |
| chr3_-_197300194 | 0.22 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr10_-_69597810 | 0.22 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chrX_-_15683147 | 0.22 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr1_+_70820451 | 0.22 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr16_-_4401258 | 0.22 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr15_+_80364901 | 0.22 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr7_-_143956815 | 0.21 |
ENST00000493325.1 |
OR2A7 |
olfactory receptor, family 2, subfamily A, member 7 |
| chr1_-_153283194 | 0.21 |
ENST00000290722.1 |
PGLYRP3 |
peptidoglycan recognition protein 3 |
| chr10_+_5135981 | 0.21 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr11_-_85430163 | 0.21 |
ENST00000529581.1 ENST00000533577.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr12_-_123756781 | 0.20 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr10_-_1246317 | 0.20 |
ENST00000381305.1 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr4_+_69313145 | 0.20 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr3_+_57882024 | 0.20 |
ENST00000494088.1 |
SLMAP |
sarcolemma associated protein |
| chr6_+_41604747 | 0.20 |
ENST00000419164.1 ENST00000373051.2 |
MDFI |
MyoD family inhibitor |
| chr6_+_35996859 | 0.20 |
ENST00000472333.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr6_-_33041378 | 0.20 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr8_-_87242589 | 0.20 |
ENST00000419776.2 ENST00000297524.3 |
SLC7A13 |
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr4_-_143227088 | 0.20 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr22_-_29107919 | 0.20 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr2_+_232573208 | 0.19 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr2_+_74685413 | 0.19 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr11_+_59824127 | 0.19 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr16_-_71610985 | 0.19 |
ENST00000355962.4 |
TAT |
tyrosine aminotransferase |
| chr18_-_19997878 | 0.19 |
ENST00000391403.2 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr18_+_55888767 | 0.19 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_+_1080164 | 0.18 |
ENST00000314040.1 |
AL033381.1 |
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr15_-_67813924 | 0.18 |
ENST00000559298.1 |
IQCH-AS1 |
IQCH antisense RNA 1 |
| chr11_-_102651343 | 0.18 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr7_-_122339162 | 0.18 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr8_+_118532937 | 0.18 |
ENST00000297347.3 |
MED30 |
mediator complex subunit 30 |
| chr6_+_32812568 | 0.18 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_+_41540160 | 0.18 |
ENST00000503057.1 ENST00000511496.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr2_-_130031335 | 0.18 |
ENST00000375987.3 |
AC079586.1 |
AC079586.1 |
| chr4_-_153274078 | 0.18 |
ENST00000263981.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr2_+_113931513 | 0.18 |
ENST00000245796.6 ENST00000441564.3 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
| chr2_+_73114489 | 0.17 |
ENST00000234454.5 |
SPR |
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) |
| chr5_-_35938674 | 0.17 |
ENST00000397366.1 ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL |
calcyphosine-like |
| chr18_-_33647487 | 0.17 |
ENST00000590898.1 ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
| chr3_+_66271410 | 0.17 |
ENST00000336733.6 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr11_-_114466477 | 0.17 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr22_+_29168652 | 0.17 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr8_+_104831554 | 0.17 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr10_+_6625605 | 0.17 |
ENST00000414894.1 ENST00000449648.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr19_+_30433110 | 0.17 |
ENST00000542441.2 ENST00000392271.1 |
URI1 |
URI1, prefoldin-like chaperone |
| chr3_+_66271489 | 0.17 |
ENST00000536651.1 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr11_-_13517565 | 0.17 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr22_+_39077947 | 0.16 |
ENST00000216034.4 |
TOMM22 |
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr4_+_41614720 | 0.16 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr3_-_190167571 | 0.16 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr4_+_41614909 | 0.16 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr4_-_87028478 | 0.16 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr10_-_75226166 | 0.16 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_140174429 | 0.16 |
ENST00000520672.2 ENST00000378132.1 ENST00000526136.1 |
PCDHA2 |
protocadherin alpha 2 |
| chr8_+_52730143 | 0.16 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr2_+_232573222 | 0.16 |
ENST00000341369.7 ENST00000409683.1 |
PTMA |
prothymosin, alpha |
| chr6_-_10435032 | 0.16 |
ENST00000491317.1 ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518 |
long intergenic non-protein coding RNA 518 |
| chr11_+_59824060 | 0.16 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr3_-_119278376 | 0.16 |
ENST00000478182.1 |
CD80 |
CD80 molecule |
| chr14_+_22675388 | 0.15 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr22_+_26879817 | 0.15 |
ENST00000215917.7 |
SRRD |
SRR1 domain containing |
| chr11_+_5509915 | 0.15 |
ENST00000322641.5 |
OR52D1 |
olfactory receptor, family 52, subfamily D, member 1 |
| chr19_-_39466396 | 0.15 |
ENST00000292852.4 |
FBXO17 |
F-box protein 17 |
| chr11_-_85430088 | 0.15 |
ENST00000533057.1 ENST00000533892.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr4_+_119200215 | 0.15 |
ENST00000602573.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr4_-_159080806 | 0.15 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr12_-_49463753 | 0.15 |
ENST00000301068.6 |
RHEBL1 |
Ras homolog enriched in brain like 1 |
| chr19_-_33360647 | 0.15 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr12_-_95044309 | 0.15 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr4_-_48082192 | 0.15 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr11_+_127140956 | 0.14 |
ENST00000608214.1 |
RP11-480C22.1 |
RP11-480C22.1 |
| chr3_-_116163830 | 0.14 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr4_-_123377880 | 0.14 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr4_+_147096837 | 0.14 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_1727755 | 0.14 |
ENST00000424383.2 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr8_+_99956662 | 0.14 |
ENST00000523368.1 ENST00000297565.4 ENST00000435298.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr9_+_12775011 | 0.14 |
ENST00000319264.3 |
LURAP1L |
leucine rich adaptor protein 1-like |
| chr6_-_137365402 | 0.14 |
ENST00000541547.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr2_-_203103185 | 0.14 |
ENST00000409205.1 |
SUMO1 |
small ubiquitin-like modifier 1 |
| chr2_+_234160217 | 0.14 |
ENST00000392017.4 ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr6_-_32498046 | 0.13 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr12_-_91398796 | 0.13 |
ENST00000261172.3 ENST00000551767.1 |
EPYC |
epiphycan |
| chr4_-_87281224 | 0.13 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr5_+_147582348 | 0.13 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr15_-_42076229 | 0.13 |
ENST00000597767.1 |
AC073657.1 |
Uncharacterized protein |
| chr10_-_62493223 | 0.13 |
ENST00000373827.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_+_100897130 | 0.13 |
ENST00000551379.1 ENST00000188403.7 ENST00000551184.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_47983519 | 0.13 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr8_-_102216925 | 0.13 |
ENST00000517844.1 |
ZNF706 |
zinc finger protein 706 |
| chr15_+_67814008 | 0.13 |
ENST00000557807.1 |
C15orf61 |
chromosome 15 open reading frame 61 |
| chrX_+_103031758 | 0.12 |
ENST00000303958.2 ENST00000361621.2 |
PLP1 |
proteolipid protein 1 |
| chr11_-_62609281 | 0.12 |
ENST00000525239.1 ENST00000538098.2 |
WDR74 |
WD repeat domain 74 |
| chr10_-_69597915 | 0.12 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_186430222 | 0.12 |
ENST00000391997.2 |
PDC |
phosducin |
| chr13_+_32313658 | 0.12 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr6_-_40555176 | 0.12 |
ENST00000338305.6 |
LRFN2 |
leucine rich repeat and fibronectin type III domain containing 2 |
| chr4_+_155484103 | 0.12 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr10_-_69597828 | 0.12 |
ENST00000339758.7 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr4_-_143226979 | 0.12 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr17_+_7788104 | 0.12 |
ENST00000380358.4 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr10_+_106113515 | 0.12 |
ENST00000369704.3 ENST00000312902.5 |
CCDC147 |
coiled-coil domain containing 147 |
| chr12_+_21525818 | 0.11 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr1_-_89591749 | 0.11 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chr13_-_47012325 | 0.11 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr11_+_57520715 | 0.11 |
ENST00000524630.1 ENST00000529919.1 ENST00000399039.4 ENST00000533189.1 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
| chr4_-_68749699 | 0.11 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr14_+_23654525 | 0.11 |
ENST00000399910.1 ENST00000492621.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr4_-_89951028 | 0.11 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr1_-_216978709 | 0.11 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr14_-_57277163 | 0.11 |
ENST00000555006.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr8_-_124279627 | 0.10 |
ENST00000357082.4 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
| chr6_+_138725343 | 0.10 |
ENST00000607197.1 ENST00000367697.3 |
HEBP2 |
heme binding protein 2 |
| chr1_+_66458072 | 0.10 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr14_+_67291158 | 0.10 |
ENST00000555456.1 |
GPHN |
gephyrin |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.6 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.4 | GO:0061346 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.6 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.3 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.4 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.7 | GO:0035234 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0051714 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0001080 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 1.4 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.1 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.6 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.2 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.3 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |