Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for CDX1
Z-value: 0.57
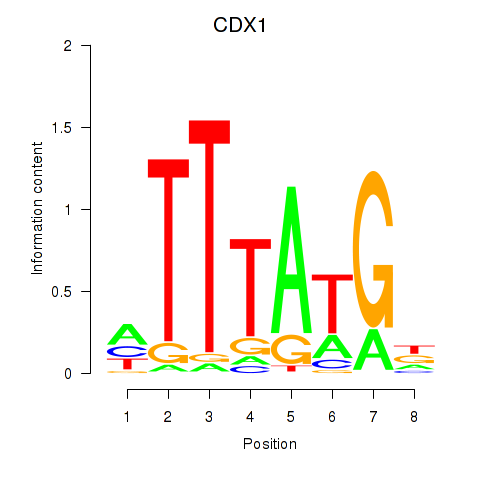
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | CDX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX1 | hg19_v2_chr5_+_149546334_149546364 | -0.60 | 1.2e-01 | Click! |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111093759 | 0.73 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr2_+_33359646 | 0.51 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.51 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr12_-_91573132 | 0.47 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr9_-_95186739 | 0.45 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr1_+_223101757 | 0.42 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr3_-_100712352 | 0.38 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr9_-_95244781 | 0.33 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr16_-_18887627 | 0.32 |
ENST00000563235.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_-_92777606 | 0.32 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr13_+_76334498 | 0.31 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr7_+_120629653 | 0.30 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr8_-_120605194 | 0.29 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_-_149095652 | 0.29 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr12_+_59989918 | 0.29 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_102826434 | 0.28 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr8_+_38586068 | 0.27 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr17_-_53809473 | 0.25 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr5_-_75919253 | 0.24 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr2_-_158345462 | 0.22 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr11_-_2158507 | 0.22 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr21_+_25801041 | 0.21 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr3_-_49314640 | 0.20 |
ENST00000436325.1 |
C3orf62 |
chromosome 3 open reading frame 62 |
| chr1_+_178310581 | 0.20 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr13_-_33780133 | 0.19 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr2_-_158300556 | 0.19 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr14_-_53331239 | 0.18 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr9_-_123239632 | 0.18 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr16_+_53241854 | 0.17 |
ENST00000565803.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr8_+_16884740 | 0.17 |
ENST00000318063.5 |
MICU3 |
mitochondrial calcium uptake family, member 3 |
| chr18_+_32455201 | 0.17 |
ENST00000590831.2 |
DTNA |
dystrobrevin, alpha |
| chr5_+_140718396 | 0.17 |
ENST00000394576.2 |
PCDHGA2 |
protocadherin gamma subfamily A, 2 |
| chr11_+_133938820 | 0.16 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr18_-_712618 | 0.16 |
ENST00000583771.1 ENST00000383578.3 ENST00000251101.7 |
ENOSF1 |
enolase superfamily member 1 |
| chr5_-_138210977 | 0.16 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr4_-_175443943 | 0.15 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr14_+_22615942 | 0.15 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr12_-_90049878 | 0.15 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_+_124125286 | 0.14 |
ENST00000368416.1 ENST00000368417.1 ENST00000546092.1 |
NKAIN2 |
Na+/K+ transporting ATPase interacting 2 |
| chr8_+_38585704 | 0.14 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr5_+_140602904 | 0.14 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr12_-_11150474 | 0.13 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr8_-_42065187 | 0.13 |
ENST00000270189.6 ENST00000352041.3 ENST00000220809.4 |
PLAT |
plasminogen activator, tissue |
| chr12_-_90049828 | 0.13 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_140739537 | 0.12 |
ENST00000522605.1 |
PCDHGB2 |
protocadherin gamma subfamily B, 2 |
| chr12_+_58013693 | 0.12 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr2_+_233527443 | 0.12 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr8_+_36641842 | 0.12 |
ENST00000523973.1 ENST00000399881.3 |
KCNU1 |
potassium channel, subfamily U, member 1 |
| chr2_-_79315112 | 0.12 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr2_-_40657397 | 0.11 |
ENST00000408028.2 ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_-_224467093 | 0.11 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr9_-_21482312 | 0.11 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr12_-_91573249 | 0.11 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr15_-_55700522 | 0.11 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr20_+_46130619 | 0.11 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr6_+_28249332 | 0.11 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr4_+_170581213 | 0.11 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr12_+_32655048 | 0.11 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_28249299 | 0.11 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr4_+_74606223 | 0.11 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr20_+_46130671 | 0.11 |
ENST00000371998.3 ENST00000371997.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr5_+_140792614 | 0.11 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr5_-_16742330 | 0.11 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr5_-_61031495 | 0.10 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr7_+_138943265 | 0.10 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr10_-_22292613 | 0.10 |
ENST00000376980.3 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr8_-_145047688 | 0.10 |
ENST00000356346.3 |
PLEC |
plectin |
| chr12_-_31744031 | 0.10 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr4_-_83765613 | 0.10 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr10_-_28270795 | 0.10 |
ENST00000545014.1 |
ARMC4 |
armadillo repeat containing 4 |
| chr8_-_42065075 | 0.10 |
ENST00000429089.2 ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT |
plasminogen activator, tissue |
| chr5_-_96518907 | 0.10 |
ENST00000508447.1 ENST00000283109.3 |
RIOK2 |
RIO kinase 2 |
| chr15_-_55700457 | 0.10 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr12_-_118796910 | 0.10 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr5_+_102201509 | 0.09 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_102201430 | 0.09 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_140588269 | 0.09 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr22_+_25595817 | 0.09 |
ENST00000215855.2 ENST00000404334.1 |
CRYBB3 |
crystallin, beta B3 |
| chr7_+_80275953 | 0.09 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr1_+_161736072 | 0.09 |
ENST00000367942.3 |
ATF6 |
activating transcription factor 6 |
| chr1_+_104159999 | 0.09 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr20_+_56964169 | 0.09 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr6_+_33168637 | 0.09 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr12_+_72079842 | 0.08 |
ENST00000266673.5 ENST00000550524.1 |
TMEM19 |
transmembrane protein 19 |
| chr14_-_106518922 | 0.08 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr6_+_33168597 | 0.08 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr5_-_78281603 | 0.08 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr10_-_81320151 | 0.08 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chrX_-_15333775 | 0.08 |
ENST00000480796.1 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr19_-_23578220 | 0.08 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr12_+_14927270 | 0.08 |
ENST00000544848.1 |
H2AFJ |
H2A histone family, member J |
| chr5_+_137419581 | 0.08 |
ENST00000506684.1 ENST00000504809.1 ENST00000398754.1 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
| chr3_+_45636219 | 0.08 |
ENST00000273317.4 |
LIMD1 |
LIM domains containing 1 |
| chr5_-_127674883 | 0.07 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr4_-_83769996 | 0.07 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr7_+_142364193 | 0.07 |
ENST00000390397.2 |
TRBV24-1 |
T cell receptor beta variable 24-1 |
| chr20_+_30795664 | 0.07 |
ENST00000375749.3 ENST00000375730.3 ENST00000539210.1 |
POFUT1 |
protein O-fucosyltransferase 1 |
| chr12_+_51318513 | 0.07 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr5_+_140571902 | 0.07 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr1_-_182921119 | 0.07 |
ENST00000423786.1 |
SHCBP1L |
SHC SH2-domain binding protein 1-like |
| chr2_+_132479948 | 0.07 |
ENST00000355171.4 |
C2orf27A |
chromosome 2 open reading frame 27A |
| chr5_-_150138061 | 0.07 |
ENST00000521533.1 ENST00000424236.1 |
DCTN4 |
dynactin 4 (p62) |
| chr1_-_152386732 | 0.07 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr5_+_140800638 | 0.07 |
ENST00000398587.2 ENST00000518882.1 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
| chr8_+_37553261 | 0.07 |
ENST00000331569.4 |
ZNF703 |
zinc finger protein 703 |
| chr15_+_22382382 | 0.07 |
ENST00000328795.4 |
OR4N4 |
olfactory receptor, family 4, subfamily N, member 4 |
| chrX_-_19689106 | 0.07 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr1_+_12290121 | 0.07 |
ENST00000358136.3 ENST00000356315.4 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr16_+_66586461 | 0.07 |
ENST00000264001.4 ENST00000351137.4 ENST00000345436.4 ENST00000362093.4 ENST00000417030.2 ENST00000527729.1 ENST00000532838.1 |
CKLF CKLF-CMTM1 |
chemokine-like factor CKLF-CMTM1 readthrough |
| chr14_+_37641012 | 0.07 |
ENST00000556667.1 |
SLC25A21-AS1 |
SLC25A21 antisense RNA 1 |
| chr18_-_712544 | 0.07 |
ENST00000340116.7 ENST00000539164.1 ENST00000580982.1 |
ENOSF1 |
enolase superfamily member 1 |
| chr4_-_100065440 | 0.07 |
ENST00000508393.1 ENST00000265512.7 |
ADH4 |
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr9_-_21335240 | 0.07 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr20_+_43595115 | 0.06 |
ENST00000372806.3 ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4 |
serine/threonine kinase 4 |
| chr6_+_84222194 | 0.06 |
ENST00000536636.1 |
PRSS35 |
protease, serine, 35 |
| chrX_-_34150428 | 0.06 |
ENST00000346193.3 |
FAM47A |
family with sequence similarity 47, member A |
| chr2_+_161993412 | 0.06 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_+_161993465 | 0.06 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr11_-_89224508 | 0.06 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chrY_-_13524717 | 0.06 |
ENST00000331172.6 |
SLC9B1P1 |
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr11_-_2182388 | 0.06 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr10_-_22292675 | 0.06 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr8_-_37797621 | 0.06 |
ENST00000524298.1 ENST00000307599.4 |
GOT1L1 |
glutamic-oxaloacetic transaminase 1-like 1 |
| chrX_+_99899180 | 0.06 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr8_+_7693993 | 0.06 |
ENST00000314265.2 |
DEFB104A |
defensin, beta 104A |
| chr18_-_31802282 | 0.06 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr1_+_12524965 | 0.06 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_+_108541608 | 0.06 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr11_-_51412448 | 0.06 |
ENST00000319760.6 |
OR4A5 |
olfactory receptor, family 4, subfamily A, member 5 |
| chr4_-_16085340 | 0.06 |
ENST00000508167.1 |
PROM1 |
prominin 1 |
| chr7_+_80267973 | 0.06 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr14_+_58894706 | 0.06 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chr21_+_31768348 | 0.06 |
ENST00000355459.2 |
KRTAP13-1 |
keratin associated protein 13-1 |
| chrX_+_36246735 | 0.06 |
ENST00000378653.3 |
CXorf30 |
chromosome X open reading frame 30 |
| chr10_+_112327425 | 0.05 |
ENST00000361804.4 |
SMC3 |
structural maintenance of chromosomes 3 |
| chr12_-_48164812 | 0.05 |
ENST00000549151.1 ENST00000548919.1 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr3_-_155394152 | 0.05 |
ENST00000494598.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr6_+_84222220 | 0.05 |
ENST00000369700.3 |
PRSS35 |
protease, serine, 35 |
| chr7_+_75024903 | 0.05 |
ENST00000323819.3 ENST00000430211.1 |
TRIM73 |
tripartite motif containing 73 |
| chr2_-_203735586 | 0.05 |
ENST00000454326.1 ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr6_-_15548591 | 0.05 |
ENST00000509674.1 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr3_+_108541545 | 0.05 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_14942283 | 0.05 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr4_-_16085314 | 0.05 |
ENST00000510224.1 |
PROM1 |
prominin 1 |
| chr14_-_92198403 | 0.05 |
ENST00000553329.1 ENST00000256343.3 |
CATSPERB |
catsper channel auxiliary subunit beta |
| chr1_-_205325850 | 0.05 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chr8_+_94752349 | 0.05 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr18_-_31802056 | 0.05 |
ENST00000538587.1 |
NOL4 |
nucleolar protein 4 |
| chrX_+_18725758 | 0.05 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr3_-_52713729 | 0.05 |
ENST00000296302.7 ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1 |
polybromo 1 |
| chr16_+_14927538 | 0.05 |
ENST00000287667.7 |
NOMO1 |
NODAL modulator 1 |
| chr2_+_46926048 | 0.05 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr5_+_108083517 | 0.05 |
ENST00000281092.4 ENST00000536402.1 |
FER |
fer (fps/fes related) tyrosine kinase |
| chr4_-_46996424 | 0.05 |
ENST00000264318.3 |
GABRA4 |
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr6_-_30658745 | 0.05 |
ENST00000376420.5 ENST00000376421.5 |
NRM |
nurim (nuclear envelope membrane protein) |
| chr15_+_55700741 | 0.05 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr17_-_46690839 | 0.05 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr10_+_97733786 | 0.05 |
ENST00000371198.2 |
CC2D2B |
coiled-coil and C2 domain containing 2B |
| chrX_+_45364633 | 0.05 |
ENST00000435394.1 ENST00000609127.1 |
RP11-245M24.1 |
RP11-245M24.1 |
| chr15_-_35047166 | 0.05 |
ENST00000290374.4 |
GJD2 |
gap junction protein, delta 2, 36kDa |
| chr14_-_81425828 | 0.05 |
ENST00000555529.1 ENST00000556042.1 ENST00000556981.1 |
CEP128 |
centrosomal protein 128kDa |
| chr3_-_168865522 | 0.05 |
ENST00000464456.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr17_-_29641104 | 0.05 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr2_-_151344172 | 0.05 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr1_-_54411240 | 0.05 |
ENST00000371378.2 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr7_-_14942944 | 0.05 |
ENST00000403951.2 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr9_-_136242909 | 0.05 |
ENST00000371991.3 ENST00000545297.1 |
SURF4 |
surfeit 4 |
| chr6_-_24489842 | 0.05 |
ENST00000230036.1 |
GPLD1 |
glycosylphosphatidylinositol specific phospholipase D1 |
| chr9_-_138391692 | 0.05 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr10_+_57358750 | 0.05 |
ENST00000512524.2 |
MTRNR2L5 |
MT-RNR2-like 5 |
| chrX_-_45629661 | 0.05 |
ENST00000602507.1 ENST00000602461.1 |
RP6-99M1.2 |
RP6-99M1.2 |
| chr5_+_129083772 | 0.05 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr5_+_95997918 | 0.05 |
ENST00000395812.2 ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST |
calpastatin |
| chrX_+_130192318 | 0.04 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr2_+_204801471 | 0.04 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chrX_-_99987088 | 0.04 |
ENST00000372981.1 ENST00000263033.5 |
SYTL4 |
synaptotagmin-like 4 |
| chr12_+_72080253 | 0.04 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chr9_-_21335356 | 0.04 |
ENST00000359039.4 |
KLHL9 |
kelch-like family member 9 |
| chr15_+_80733570 | 0.04 |
ENST00000533983.1 ENST00000527771.1 ENST00000525103.1 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr14_+_100789669 | 0.04 |
ENST00000361529.3 ENST00000557052.1 |
SLC25A47 |
solute carrier family 25, member 47 |
| chr12_-_111358372 | 0.04 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chrX_-_122756660 | 0.04 |
ENST00000441692.1 |
THOC2 |
THO complex 2 |
| chr11_+_61129456 | 0.04 |
ENST00000278826.6 |
TMEM138 |
transmembrane protein 138 |
| chr5_-_138775177 | 0.04 |
ENST00000302060.5 |
DNAJC18 |
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_+_64920543 | 0.04 |
ENST00000399438.3 ENST00000510585.2 |
TRAPPC13 CTC-534A2.2 |
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr7_+_143632259 | 0.04 |
ENST00000408955.2 |
OR2F2 |
olfactory receptor, family 2, subfamily F, member 2 |
| chr1_-_54411255 | 0.04 |
ENST00000371377.3 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr1_-_156265438 | 0.04 |
ENST00000362007.1 |
C1orf85 |
chromosome 1 open reading frame 85 |
| chr2_+_166095898 | 0.04 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_+_36065053 | 0.04 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr14_-_54955721 | 0.04 |
ENST00000554908.1 |
GMFB |
glia maturation factor, beta |
| chr7_+_106505912 | 0.04 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_154975110 | 0.04 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr4_+_159727272 | 0.04 |
ENST00000379346.3 |
FNIP2 |
folliculin interacting protein 2 |
| chr9_-_97356075 | 0.04 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr19_+_15852203 | 0.04 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr22_+_40322595 | 0.04 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr1_+_84630645 | 0.04 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_170366203 | 0.04 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr2_+_169757750 | 0.04 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.0 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |