Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
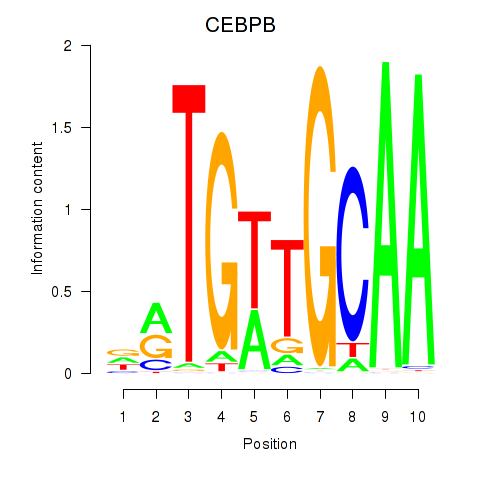
Results for CEBPB
Z-value: 0.62
Transcription factors associated with CEBPB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPB
|
ENSG00000172216.4 | CEBPB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPB | hg19_v2_chr20_+_48807351_48807384 | -0.07 | 8.7e-01 | Click! |
Activity profile of CEBPB motif
Sorted Z-values of CEBPB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 1.67 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 1.38 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr4_+_113739244 | 1.37 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr17_+_1665345 | 1.08 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_-_56236734 | 0.98 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr3_+_157154578 | 0.93 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr12_-_56236690 | 0.92 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr16_-_28550320 | 0.90 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr16_-_28550348 | 0.88 |
ENST00000324873.6 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr3_+_12392971 | 0.86 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr22_+_35776828 | 0.81 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr21_-_44495919 | 0.80 |
ENST00000398158.1 |
CBS |
cystathionine-beta-synthase |
| chr17_+_1665253 | 0.80 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr21_-_44495964 | 0.79 |
ENST00000398168.1 ENST00000398165.3 |
CBS |
cystathionine-beta-synthase |
| chr7_-_56101826 | 0.77 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr5_-_172756506 | 0.71 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr4_-_139163491 | 0.68 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr15_+_41245160 | 0.67 |
ENST00000444189.2 ENST00000446533.3 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr9_+_2621798 | 0.65 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr3_-_66551351 | 0.59 |
ENST00000273261.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr3_-_66551397 | 0.56 |
ENST00000383703.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_-_70725856 | 0.55 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr6_+_73076432 | 0.54 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr2_+_65216462 | 0.51 |
ENST00000234256.3 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_221051699 | 0.49 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr2_-_175711133 | 0.45 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr9_+_131683174 | 0.43 |
ENST00000372592.3 ENST00000428610.1 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
| chr4_+_77870960 | 0.42 |
ENST00000505788.1 ENST00000510515.1 ENST00000504637.1 |
SEPT11 |
septin 11 |
| chr10_+_95848824 | 0.40 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr1_+_28586006 | 0.40 |
ENST00000253063.3 |
SESN2 |
sestrin 2 |
| chr3_+_141105705 | 0.39 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr19_+_1105110 | 0.38 |
ENST00000587648.1 |
GPX4 |
glutathione peroxidase 4 |
| chr7_+_48211048 | 0.37 |
ENST00000435803.1 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr4_+_77870856 | 0.35 |
ENST00000264893.6 ENST00000502584.1 ENST00000510641.1 |
SEPT11 |
septin 11 |
| chr14_-_100841670 | 0.34 |
ENST00000557297.1 ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr17_-_79895097 | 0.34 |
ENST00000402252.2 ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr1_+_164528866 | 0.34 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr19_-_11347173 | 0.33 |
ENST00000587656.1 |
DOCK6 |
dedicator of cytokinesis 6 |
| chr11_+_47279504 | 0.32 |
ENST00000441012.2 ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr9_-_13175823 | 0.32 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr17_-_33390667 | 0.31 |
ENST00000378516.2 ENST00000268850.7 ENST00000394597.2 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr17_-_79895154 | 0.31 |
ENST00000405481.4 ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr1_-_44497118 | 0.30 |
ENST00000537678.1 ENST00000466926.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_+_57624085 | 0.30 |
ENST00000553474.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_50860565 | 0.28 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr6_+_31895467 | 0.28 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_+_126102292 | 0.28 |
ENST00000368357.3 |
NCOA7 |
nuclear receptor coactivator 7 |
| chrX_-_118284542 | 0.27 |
ENST00000402510.2 |
KIAA1210 |
KIAA1210 |
| chr1_+_153232160 | 0.27 |
ENST00000368742.3 |
LOR |
loricrin |
| chr12_-_25101920 | 0.27 |
ENST00000539780.1 ENST00000546285.1 ENST00000342945.5 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_25102252 | 0.26 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_96390108 | 0.26 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr17_+_29248953 | 0.25 |
ENST00000581285.1 |
ADAP2 |
ArfGAP with dual PH domains 2 |
| chr1_-_203198790 | 0.25 |
ENST00000367229.1 ENST00000255427.3 ENST00000535569.1 |
CHIT1 |
chitinase 1 (chitotriosidase) |
| chr2_+_46769798 | 0.24 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr14_-_100841930 | 0.23 |
ENST00000555031.1 ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chrX_-_43832711 | 0.23 |
ENST00000378062.5 |
NDP |
Norrie disease (pseudoglioma) |
| chr1_-_38412683 | 0.22 |
ENST00000373024.3 ENST00000373023.2 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
| chr11_-_33913708 | 0.22 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr3_+_141106458 | 0.21 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr7_+_114562616 | 0.21 |
ENST00000448022.1 |
MDFIC |
MyoD family inhibitor domain containing |
| chr12_+_57624119 | 0.21 |
ENST00000555773.1 ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr3_-_49722523 | 0.21 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_114562172 | 0.21 |
ENST00000393486.1 ENST00000257724.3 |
MDFIC |
MyoD family inhibitor domain containing |
| chr19_-_44123734 | 0.21 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr11_+_73019282 | 0.20 |
ENST00000263674.3 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr9_+_2015335 | 0.19 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_-_154493791 | 0.19 |
ENST00000369454.3 |
RAB39B |
RAB39B, member RAS oncogene family |
| chr2_-_43453734 | 0.18 |
ENST00000282388.3 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
| chr1_-_196577489 | 0.18 |
ENST00000609185.1 ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2 |
potassium channel, subfamily T, member 2 |
| chr4_+_71063641 | 0.18 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr1_-_108231101 | 0.18 |
ENST00000544443.1 ENST00000415432.2 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr1_-_149908710 | 0.17 |
ENST00000439741.2 ENST00000361405.6 ENST00000406732.3 |
MTMR11 |
myotubularin related protein 11 |
| chr1_+_154244987 | 0.17 |
ENST00000328703.7 ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1 |
HCLS1 associated protein X-1 |
| chr1_-_149908217 | 0.17 |
ENST00000369140.3 |
MTMR11 |
myotubularin related protein 11 |
| chr1_-_85155939 | 0.17 |
ENST00000603677.1 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr12_+_56390964 | 0.17 |
ENST00000356124.4 ENST00000266971.3 ENST00000394115.2 ENST00000547586.1 ENST00000552258.1 ENST00000548274.1 ENST00000546833.1 |
SUOX |
sulfite oxidase |
| chr12_+_57623869 | 0.16 |
ENST00000414700.3 ENST00000557703.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_-_132113559 | 0.16 |
ENST00000448933.1 |
SEPT8 |
septin 8 |
| chr1_-_150602035 | 0.16 |
ENST00000503241.1 ENST00000369016.4 ENST00000339643.5 ENST00000271690.8 ENST00000356527.5 ENST00000362052.7 ENST00000503345.1 ENST00000369014.5 ENST00000369009.3 |
ENSA |
endosulfine alpha |
| chr19_-_44124019 | 0.16 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr6_+_31895254 | 0.16 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chrX_-_106243451 | 0.16 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr1_-_23857698 | 0.16 |
ENST00000361729.2 |
E2F2 |
E2F transcription factor 2 |
| chr1_-_150601588 | 0.15 |
ENST00000513281.1 ENST00000361631.5 ENST00000361532.5 |
ENSA |
endosulfine alpha |
| chr11_-_3078616 | 0.15 |
ENST00000401769.3 ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS |
cysteinyl-tRNA synthetase |
| chr20_+_56136136 | 0.15 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_+_57849048 | 0.15 |
ENST00000266646.2 |
INHBE |
inhibin, beta E |
| chr11_-_3078838 | 0.15 |
ENST00000397111.5 |
CARS |
cysteinyl-tRNA synthetase |
| chr10_+_111765562 | 0.15 |
ENST00000360162.3 |
ADD3 |
adducin 3 (gamma) |
| chr1_+_52682052 | 0.15 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr2_+_211421262 | 0.14 |
ENST00000233072.5 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr1_-_44497024 | 0.14 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_+_57623477 | 0.14 |
ENST00000557487.1 ENST00000555634.1 ENST00000556689.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr16_-_15736881 | 0.14 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr5_+_139505520 | 0.13 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr6_+_31895480 | 0.13 |
ENST00000418949.2 ENST00000383177.3 ENST00000477310.1 |
C2 CFB |
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr2_-_220174166 | 0.13 |
ENST00000409251.3 ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr19_+_33865218 | 0.13 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr17_+_44588877 | 0.13 |
ENST00000576629.1 |
LRRC37A2 |
leucine rich repeat containing 37, member A2 |
| chr6_-_32812420 | 0.12 |
ENST00000374881.2 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr20_+_39969519 | 0.12 |
ENST00000373257.3 |
LPIN3 |
lipin 3 |
| chr3_-_126327398 | 0.12 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr20_+_361890 | 0.12 |
ENST00000449710.1 ENST00000422053.2 |
TRIB3 |
tribbles pseudokinase 3 |
| chrX_-_109683446 | 0.12 |
ENST00000372057.1 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr19_-_51869592 | 0.12 |
ENST00000596253.1 ENST00000309244.4 |
ETFB |
electron-transfer-flavoprotein, beta polypeptide |
| chr1_+_50574585 | 0.12 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr1_+_52521928 | 0.12 |
ENST00000489308.2 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr15_+_89182178 | 0.12 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr19_-_36236292 | 0.11 |
ENST00000378975.3 ENST00000412391.2 ENST00000292879.5 |
U2AF1L4 |
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr2_+_227700652 | 0.11 |
ENST00000341329.3 ENST00000392062.2 ENST00000437454.1 ENST00000443477.1 ENST00000423616.1 ENST00000448992.1 |
RHBDD1 |
rhomboid domain containing 1 |
| chr15_+_71145578 | 0.11 |
ENST00000544974.2 ENST00000558546.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr1_+_52521797 | 0.11 |
ENST00000313334.8 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr5_-_179047881 | 0.11 |
ENST00000521173.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr22_+_39052632 | 0.11 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr2_-_160654745 | 0.11 |
ENST00000259053.4 ENST00000429078.2 |
CD302 |
CD302 molecule |
| chr10_+_45495898 | 0.11 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr15_+_89182156 | 0.11 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr6_+_32812568 | 0.11 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr17_-_77813186 | 0.11 |
ENST00000448310.1 ENST00000269397.4 |
CBX4 |
chromobox homolog 4 |
| chr16_+_3162557 | 0.11 |
ENST00000382192.3 ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205 |
zinc finger protein 205 |
| chr2_+_121010413 | 0.11 |
ENST00000404963.3 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
| chr1_+_53480598 | 0.11 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr3_+_141105235 | 0.10 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr2_+_187350973 | 0.10 |
ENST00000544130.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr20_-_2821756 | 0.10 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr8_+_38831683 | 0.10 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr1_-_20250110 | 0.09 |
ENST00000375116.3 |
PLA2G2E |
phospholipase A2, group IIE |
| chr4_+_86396321 | 0.09 |
ENST00000503995.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr20_-_2821271 | 0.09 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chr12_-_57914275 | 0.09 |
ENST00000547303.1 ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3 |
DNA-damage-inducible transcript 3 |
| chr2_+_28618532 | 0.09 |
ENST00000545753.1 |
FOSL2 |
FOS-like antigen 2 |
| chr1_+_53527854 | 0.09 |
ENST00000371500.3 ENST00000395871.2 ENST00000312553.5 |
PODN |
podocan |
| chr20_+_8112824 | 0.09 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr1_-_220219775 | 0.08 |
ENST00000609181.1 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr15_-_64665911 | 0.08 |
ENST00000606793.1 ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1 |
Uncharacterized protein |
| chr5_-_149792295 | 0.08 |
ENST00000518797.1 ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr15_+_96869165 | 0.08 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_99127225 | 0.08 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr17_-_38978847 | 0.07 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr5_+_150020214 | 0.07 |
ENST00000307662.4 |
SYNPO |
synaptopodin |
| chr3_-_71179699 | 0.07 |
ENST00000497355.1 |
FOXP1 |
forkhead box P1 |
| chr12_+_6644443 | 0.07 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr20_-_32700075 | 0.07 |
ENST00000374980.2 |
EIF2S2 |
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| chr1_+_212782012 | 0.07 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr22_-_30901637 | 0.07 |
ENST00000381982.3 ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4 |
SEC14-like 4 (S. cerevisiae) |
| chr20_+_361261 | 0.07 |
ENST00000217233.3 |
TRIB3 |
tribbles pseudokinase 3 |
| chr7_+_99006232 | 0.07 |
ENST00000403633.2 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr1_-_207143802 | 0.07 |
ENST00000324852.4 ENST00000400962.3 |
FCAMR |
Fc receptor, IgA, IgM, high affinity |
| chr2_-_179567206 | 0.06 |
ENST00000414766.1 |
TTN |
titin |
| chr11_-_12030629 | 0.06 |
ENST00000396505.2 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
| chr13_+_77522632 | 0.06 |
ENST00000377462.1 |
IRG1 |
immunoresponsive 1 homolog (mouse) |
| chr1_+_67395922 | 0.06 |
ENST00000401042.3 ENST00000355356.3 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
| chrX_-_80457385 | 0.06 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr3_+_99833755 | 0.06 |
ENST00000489081.1 |
CMSS1 |
cms1 ribosomal small subunit homolog (yeast) |
| chr22_+_41258250 | 0.06 |
ENST00000544094.1 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr15_-_61521495 | 0.06 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr6_+_36238237 | 0.06 |
ENST00000457797.1 ENST00000394571.2 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
| chr16_+_1359511 | 0.06 |
ENST00000397514.3 ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chr6_+_31939608 | 0.06 |
ENST00000375331.2 ENST00000375333.2 |
STK19 |
serine/threonine kinase 19 |
| chr4_+_78978724 | 0.06 |
ENST00000325942.6 ENST00000264895.6 ENST00000264899.6 |
FRAS1 |
Fraser syndrome 1 |
| chr12_+_93096619 | 0.05 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr3_-_48057890 | 0.05 |
ENST00000434267.1 |
MAP4 |
microtubule-associated protein 4 |
| chr12_+_93096759 | 0.05 |
ENST00000544406.2 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr11_-_12030905 | 0.05 |
ENST00000326932.4 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_+_136343820 | 0.05 |
ENST00000410054.1 |
R3HDM1 |
R3H domain containing 1 |
| chr3_+_46395219 | 0.05 |
ENST00000445132.2 ENST00000292301.4 |
CCR2 |
chemokine (C-C motif) receptor 2 |
| chr3_-_99833333 | 0.05 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr11_-_102595681 | 0.05 |
ENST00000236826.3 |
MMP8 |
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr16_-_70323422 | 0.05 |
ENST00000261772.8 |
AARS |
alanyl-tRNA synthetase |
| chr12_+_123868320 | 0.05 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr12_-_96390063 | 0.05 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr4_+_86396265 | 0.05 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr20_+_36974759 | 0.04 |
ENST00000217407.2 |
LBP |
lipopolysaccharide binding protein |
| chr16_+_72088376 | 0.04 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr9_+_137218362 | 0.04 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr7_-_76829125 | 0.04 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr17_+_43239231 | 0.04 |
ENST00000591576.1 ENST00000591070.1 ENST00000592695.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chrX_-_70288234 | 0.04 |
ENST00000276105.3 ENST00000374274.3 |
SNX12 |
sorting nexin 12 |
| chr15_+_89181974 | 0.04 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr9_-_95055956 | 0.04 |
ENST00000375629.3 ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS |
isoleucyl-tRNA synthetase |
| chr6_-_31939734 | 0.04 |
ENST00000375356.3 |
DXO |
decapping exoribonuclease |
| chr1_+_29213678 | 0.04 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr11_-_8680383 | 0.04 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr7_+_28725585 | 0.04 |
ENST00000396298.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr7_+_99006550 | 0.04 |
ENST00000222969.5 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr4_-_38858428 | 0.03 |
ENST00000436693.2 ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6 TLR1 |
toll-like receptor 6 toll-like receptor 1 |
| chr22_+_40573921 | 0.03 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr19_+_11350278 | 0.03 |
ENST00000252453.8 |
C19orf80 |
chromosome 19 open reading frame 80 |
| chrX_+_13587712 | 0.03 |
ENST00000361306.1 ENST00000380602.3 |
EGFL6 |
EGF-like-domain, multiple 6 |
| chr5_-_145562147 | 0.03 |
ENST00000545646.1 ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS |
leucyl-tRNA synthetase |
| chr15_+_62359175 | 0.03 |
ENST00000355522.5 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
| chr16_-_67597789 | 0.03 |
ENST00000605277.1 |
CTD-2012K14.6 |
CTD-2012K14.6 |
| chr19_-_54824344 | 0.03 |
ENST00000346508.3 ENST00000446712.3 ENST00000432233.3 ENST00000301219.3 |
LILRA5 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr16_+_58426296 | 0.03 |
ENST00000426538.2 ENST00000328514.7 ENST00000318129.5 |
GINS3 |
GINS complex subunit 3 (Psf3 homolog) |
| chr9_-_34691201 | 0.03 |
ENST00000378800.3 ENST00000311925.2 |
CCL19 |
chemokine (C-C motif) ligand 19 |
| chr5_+_167956121 | 0.03 |
ENST00000338333.4 |
FBLL1 |
fibrillarin-like 1 |
| chr14_-_25045446 | 0.03 |
ENST00000216336.2 |
CTSG |
cathepsin G |
| chr12_+_10124001 | 0.03 |
ENST00000396507.3 ENST00000304361.4 ENST00000434319.2 |
CLEC12A |
C-type lectin domain family 12, member A |
| chr4_-_134070250 | 0.02 |
ENST00000505289.1 ENST00000509715.1 |
RP11-9G1.3 |
RP11-9G1.3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.5 | 1.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 0.8 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.3 | 3.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.9 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 1.9 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.9 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.6 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.0 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.3 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.5 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.5 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |