Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for CLOCK
Z-value: 0.44
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
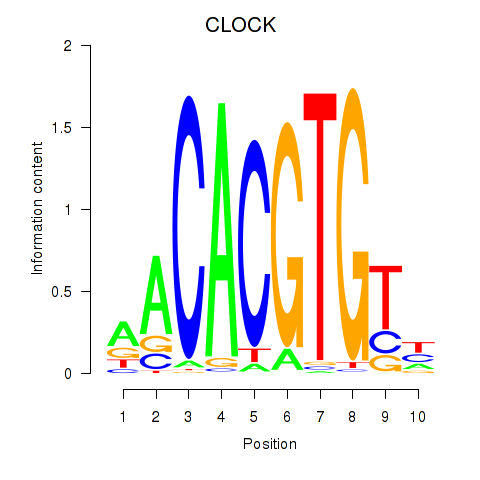
CLOCK
|
ENSG00000134852.10 | CLOCK |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_110427983 | 0.49 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr11_+_72929402 | 0.47 |
ENST00000393596.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_72929319 | 0.47 |
ENST00000393597.2 ENST00000311131.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr2_-_31361543 | 0.45 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr15_+_40531243 | 0.44 |
ENST00000558055.1 ENST00000455577.2 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr15_+_40531621 | 0.43 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_17721230 | 0.41 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chr18_+_33877654 | 0.41 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr4_+_41258786 | 0.41 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr14_+_61789382 | 0.39 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr22_+_40390930 | 0.39 |
ENST00000333407.6 |
FAM83F |
family with sequence similarity 83, member F |
| chr17_-_4458616 | 0.38 |
ENST00000381556.2 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chr1_+_62439037 | 0.38 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr2_+_17721920 | 0.37 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chrX_-_106449656 | 0.36 |
ENST00000372466.4 ENST00000421752.1 ENST00000372461.3 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
| chr4_+_40198527 | 0.36 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr2_+_131769256 | 0.35 |
ENST00000355771.3 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr17_+_55163075 | 0.33 |
ENST00000571629.1 ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr17_-_38657849 | 0.32 |
ENST00000254051.6 |
TNS4 |
tensin 4 |
| chr6_+_41888926 | 0.32 |
ENST00000230340.4 |
BYSL |
bystin-like |
| chr3_-_50329835 | 0.32 |
ENST00000429673.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chr21_+_45527171 | 0.31 |
ENST00000291576.7 ENST00000456705.1 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
| chr3_-_123411191 | 0.28 |
ENST00000354792.5 ENST00000508240.1 |
MYLK |
myosin light chain kinase |
| chr17_-_2614927 | 0.28 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr3_+_184032919 | 0.26 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr12_-_25101920 | 0.25 |
ENST00000539780.1 ENST00000546285.1 ENST00000342945.5 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr12_+_101673872 | 0.24 |
ENST00000261637.4 |
UTP20 |
UTP20, small subunit (SSU) processome component, homolog (yeast) |
| chr17_-_73285293 | 0.24 |
ENST00000582778.1 ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr5_+_149737202 | 0.24 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr7_+_26191809 | 0.23 |
ENST00000056233.3 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
| chr5_+_176784837 | 0.23 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr20_-_6103666 | 0.23 |
ENST00000536936.1 |
FERMT1 |
fermitin family member 1 |
| chr19_+_29493456 | 0.23 |
ENST00000591143.1 ENST00000592653.1 |
CTD-2081K17.2 |
CTD-2081K17.2 |
| chr11_-_94227029 | 0.23 |
ENST00000323977.3 ENST00000536754.1 ENST00000323929.3 |
MRE11A |
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr1_+_11333245 | 0.22 |
ENST00000376810.5 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
| chr12_-_6740802 | 0.22 |
ENST00000431922.1 |
LPAR5 |
lysophosphatidic acid receptor 5 |
| chr12_-_25102252 | 0.22 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr11_+_75479850 | 0.22 |
ENST00000376262.3 ENST00000604733.1 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
| chr3_-_50329990 | 0.22 |
ENST00000417626.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chr4_-_103266355 | 0.22 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr17_+_30813576 | 0.21 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr11_+_75479763 | 0.21 |
ENST00000228027.7 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
| chr7_-_122339162 | 0.21 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr10_+_70715884 | 0.21 |
ENST00000354185.4 |
DDX21 |
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr10_-_6019552 | 0.20 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr4_-_123542224 | 0.20 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr4_+_95679072 | 0.20 |
ENST00000515059.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr2_+_62423242 | 0.20 |
ENST00000301998.4 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr19_+_1407733 | 0.20 |
ENST00000592453.1 |
DAZAP1 |
DAZ associated protein 1 |
| chr17_-_655489 | 0.20 |
ENST00000319004.5 |
GEMIN4 |
gem (nuclear organelle) associated protein 4 |
| chr17_-_41623716 | 0.20 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr10_-_120925054 | 0.20 |
ENST00000419372.1 ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4 |
sideroflexin 4 |
| chr9_-_139137648 | 0.19 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr6_-_39197226 | 0.19 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr12_-_31478428 | 0.19 |
ENST00000543615.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr4_-_54232144 | 0.19 |
ENST00000388940.4 ENST00000503450.1 ENST00000401642.3 |
SCFD2 |
sec1 family domain containing 2 |
| chr7_-_92465868 | 0.18 |
ENST00000424848.2 |
CDK6 |
cyclin-dependent kinase 6 |
| chr3_+_184032419 | 0.18 |
ENST00000352767.3 ENST00000427141.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr9_+_706842 | 0.18 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr3_+_46283916 | 0.18 |
ENST00000395940.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr6_-_31763721 | 0.18 |
ENST00000375663.3 |
VARS |
valyl-tRNA synthetase |
| chr1_+_150254936 | 0.18 |
ENST00000447007.1 ENST00000369095.1 ENST00000369094.1 |
C1orf51 |
chromosome 1 open reading frame 51 |
| chr2_+_118572226 | 0.18 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr6_+_12007963 | 0.18 |
ENST00000607445.1 |
RP11-456H18.2 |
RP11-456H18.2 |
| chr5_+_70883178 | 0.18 |
ENST00000323375.8 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr6_+_44187242 | 0.18 |
ENST00000393844.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr12_-_13529642 | 0.17 |
ENST00000318426.2 |
C12orf36 |
chromosome 12 open reading frame 36 |
| chr22_+_18593446 | 0.17 |
ENST00000316027.6 |
TUBA8 |
tubulin, alpha 8 |
| chr4_-_4291861 | 0.17 |
ENST00000343470.4 |
LYAR |
Ly1 antibody reactive |
| chr4_-_4291748 | 0.17 |
ENST00000452476.1 |
LYAR |
Ly1 antibody reactive |
| chr3_+_183967409 | 0.17 |
ENST00000324557.4 ENST00000402825.3 |
ECE2 |
endothelin converting enzyme 2 |
| chr6_-_43496605 | 0.17 |
ENST00000455285.2 |
XPO5 |
exportin 5 |
| chr17_-_40333099 | 0.16 |
ENST00000607371.1 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr5_+_70883117 | 0.16 |
ENST00000340941.6 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr9_-_34637718 | 0.16 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr1_+_173793777 | 0.16 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr8_-_103424916 | 0.16 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr12_-_113623252 | 0.16 |
ENST00000314045.7 ENST00000306014.5 |
DDX54 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr14_+_96722539 | 0.16 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr13_-_44735393 | 0.16 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr4_-_4291761 | 0.16 |
ENST00000513174.1 |
LYAR |
Ly1 antibody reactive |
| chr8_-_103425047 | 0.15 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr19_+_34895289 | 0.15 |
ENST00000246535.3 |
PDCD2L |
programmed cell death 2-like |
| chr1_-_15911510 | 0.15 |
ENST00000375826.3 |
AGMAT |
agmatine ureohydrolase (agmatinase) |
| chr12_+_81110684 | 0.15 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr1_+_173793641 | 0.15 |
ENST00000361951.4 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr5_+_135394840 | 0.15 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr8_-_103424986 | 0.15 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr9_-_124976185 | 0.15 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr6_+_44191507 | 0.15 |
ENST00000371724.1 ENST00000371713.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr5_+_34656331 | 0.15 |
ENST00000265109.3 |
RAI14 |
retinoic acid induced 14 |
| chr11_-_118972575 | 0.15 |
ENST00000432443.2 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr6_-_30712313 | 0.15 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr6_+_44187334 | 0.14 |
ENST00000313248.7 ENST00000427851.2 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr6_-_90529418 | 0.14 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr3_+_184033135 | 0.14 |
ENST00000424196.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr12_-_52604607 | 0.14 |
ENST00000551894.1 ENST00000553017.1 |
C12orf80 |
chromosome 12 open reading frame 80 |
| chr6_+_44191290 | 0.14 |
ENST00000371755.3 ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr6_-_47277634 | 0.14 |
ENST00000296861.2 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
| chr1_-_111991850 | 0.14 |
ENST00000411751.2 |
WDR77 |
WD repeat domain 77 |
| chr6_-_143771799 | 0.14 |
ENST00000237283.8 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
| chr2_+_68384976 | 0.14 |
ENST00000263657.2 |
PNO1 |
partner of NOB1 homolog (S. cerevisiae) |
| chr17_-_76836963 | 0.14 |
ENST00000312010.6 |
USP36 |
ubiquitin specific peptidase 36 |
| chrX_+_16804544 | 0.13 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr3_+_102153859 | 0.13 |
ENST00000306176.1 ENST00000466937.1 |
ZPLD1 |
zona pellucida-like domain containing 1 |
| chr1_-_11986442 | 0.13 |
ENST00000376572.3 ENST00000376576.3 |
KIAA2013 |
KIAA2013 |
| chr1_-_216978709 | 0.13 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_120839412 | 0.13 |
ENST00000355228.4 |
FAM72B |
family with sequence similarity 72, member B |
| chr7_-_148725733 | 0.13 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr15_+_68924327 | 0.13 |
ENST00000543950.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chrX_+_54556633 | 0.13 |
ENST00000336470.4 ENST00000360845.2 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr2_+_32853093 | 0.13 |
ENST00000448773.1 ENST00000317907.4 |
TTC27 |
tetratricopeptide repeat domain 27 |
| chr1_-_11120057 | 0.13 |
ENST00000376957.2 |
SRM |
spermidine synthase |
| chr3_+_118866222 | 0.13 |
ENST00000490594.1 |
RP11-484M3.5 |
Uncharacterized protein |
| chr17_-_9694614 | 0.13 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr17_-_40075219 | 0.13 |
ENST00000537919.1 ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY |
ATP citrate lyase |
| chr14_+_77924373 | 0.13 |
ENST00000216479.3 ENST00000535854.2 ENST00000555517.1 |
AHSA1 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr12_+_6309963 | 0.12 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr4_-_103266626 | 0.12 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr22_+_21996549 | 0.12 |
ENST00000248958.4 |
SDF2L1 |
stromal cell-derived factor 2-like 1 |
| chr2_+_173420697 | 0.12 |
ENST00000282077.3 ENST00000392571.2 ENST00000410055.1 |
PDK1 |
pyruvate dehydrogenase kinase, isozyme 1 |
| chr22_-_44258360 | 0.12 |
ENST00000330884.4 ENST00000249130.5 |
SULT4A1 |
sulfotransferase family 4A, member 1 |
| chr19_+_1071203 | 0.12 |
ENST00000543365.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr5_+_34656569 | 0.12 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr16_+_2867164 | 0.12 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr4_-_1400119 | 0.12 |
ENST00000422806.1 |
NKX1-1 |
NK1 homeobox 1 |
| chr11_-_94226964 | 0.12 |
ENST00000538923.1 ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A |
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr6_-_154677900 | 0.12 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr14_+_78266436 | 0.12 |
ENST00000557501.1 ENST00000341211.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr1_-_113498616 | 0.12 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr14_+_96722152 | 0.12 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr8_+_109455845 | 0.12 |
ENST00000220853.3 |
EMC2 |
ER membrane protein complex subunit 2 |
| chr8_+_67341239 | 0.12 |
ENST00000320270.2 |
RRS1 |
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr3_+_1134260 | 0.12 |
ENST00000446702.2 ENST00000539053.1 ENST00000350110.2 |
CNTN6 |
contactin 6 |
| chr17_+_48450575 | 0.12 |
ENST00000338165.4 ENST00000393271.2 ENST00000511519.2 |
EME1 |
essential meiotic structure-specific endonuclease 1 |
| chr1_+_20617362 | 0.12 |
ENST00000375079.2 ENST00000289815.8 ENST00000375083.4 ENST00000289825.4 |
VWA5B1 |
von Willebrand factor A domain containing 5B1 |
| chr10_+_70939983 | 0.11 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr3_+_57741957 | 0.11 |
ENST00000295951.3 |
SLMAP |
sarcolemma associated protein |
| chr10_-_5708515 | 0.11 |
ENST00000357700.6 |
ASB13 |
ankyrin repeat and SOCS box containing 13 |
| chr6_-_167797887 | 0.11 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr21_-_44299626 | 0.11 |
ENST00000330317.2 ENST00000398208.2 |
WDR4 |
WD repeat domain 4 |
| chr8_-_144442094 | 0.11 |
ENST00000521193.1 ENST00000520950.1 |
TOP1MT |
topoisomerase (DNA) I, mitochondrial |
| chr15_-_68521996 | 0.11 |
ENST00000418702.2 ENST00000565471.1 ENST00000564752.1 ENST00000566347.1 ENST00000249806.5 ENST00000562767.1 |
CLN6 RP11-315D16.2 |
ceroid-lipofuscinosis, neuronal 6, late infantile, variant Uncharacterized protein |
| chr21_-_46330545 | 0.11 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr9_-_34637806 | 0.11 |
ENST00000477726.1 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr12_-_114404111 | 0.11 |
ENST00000545145.2 ENST00000392561.3 ENST00000261741.5 |
RBM19 |
RNA binding motif protein 19 |
| chr6_+_12007897 | 0.11 |
ENST00000437559.1 |
RP11-456H18.2 |
RP11-456H18.2 |
| chr6_+_7107999 | 0.11 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr17_-_5372271 | 0.11 |
ENST00000225296.3 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr3_+_124449213 | 0.11 |
ENST00000232607.2 ENST00000536109.1 ENST00000538242.1 ENST00000413078.2 |
UMPS |
uridine monophosphate synthetase |
| chr1_-_205744205 | 0.11 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr19_+_50180317 | 0.11 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr17_+_35306175 | 0.11 |
ENST00000225402.5 |
AATF |
apoptosis antagonizing transcription factor |
| chr15_-_101835414 | 0.11 |
ENST00000254193.6 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr3_-_132378897 | 0.11 |
ENST00000545291.1 |
ACAD11 |
acyl-CoA dehydrogenase family, member 11 |
| chr18_-_33077868 | 0.11 |
ENST00000590757.1 ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C RP11-322E11.6 |
INO80 complex subunit C Uncharacterized protein |
| chr9_+_130565487 | 0.10 |
ENST00000373225.3 ENST00000431857.1 |
FPGS |
folylpolyglutamate synthase |
| chr10_+_103892787 | 0.10 |
ENST00000278070.2 ENST00000413464.2 |
PPRC1 |
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr2_-_232329186 | 0.10 |
ENST00000322723.4 |
NCL |
nucleolin |
| chr9_-_2844058 | 0.10 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr11_+_76494253 | 0.10 |
ENST00000333090.4 |
TSKU |
tsukushi, small leucine rich proteoglycan |
| chr19_+_10765003 | 0.10 |
ENST00000407004.3 ENST00000589998.1 ENST00000589600.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr11_-_47870091 | 0.10 |
ENST00000526870.1 |
NUP160 |
nucleoporin 160kDa |
| chr19_-_41903161 | 0.10 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr18_-_11670159 | 0.10 |
ENST00000561598.1 |
RP11-677O4.2 |
RP11-677O4.2 |
| chr17_-_40333150 | 0.10 |
ENST00000264661.3 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr15_-_82555000 | 0.10 |
ENST00000557844.1 ENST00000359445.3 ENST00000268206.7 |
EFTUD1 |
elongation factor Tu GTP binding domain containing 1 |
| chr11_-_61684962 | 0.10 |
ENST00000394836.2 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr11_-_125366089 | 0.10 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr22_-_30987837 | 0.10 |
ENST00000335214.6 |
PES1 |
pescadillo ribosomal biogenesis factor 1 |
| chr2_-_71454185 | 0.10 |
ENST00000244221.8 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
| chr7_+_116312411 | 0.10 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr22_-_30987849 | 0.10 |
ENST00000402284.3 ENST00000354694.7 |
PES1 |
pescadillo ribosomal biogenesis factor 1 |
| chr9_+_130565147 | 0.10 |
ENST00000373247.2 ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS |
folylpolyglutamate synthase |
| chr10_-_6019455 | 0.10 |
ENST00000530685.1 ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr1_-_111991908 | 0.10 |
ENST00000235090.5 |
WDR77 |
WD repeat domain 77 |
| chr3_-_122512619 | 0.10 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chr19_+_1067271 | 0.10 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr3_-_71834318 | 0.10 |
ENST00000353065.3 |
PROK2 |
prokineticin 2 |
| chr21_+_45209394 | 0.10 |
ENST00000497547.1 |
RRP1 |
ribosomal RNA processing 1 |
| chr17_+_72427477 | 0.10 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr4_-_57301748 | 0.10 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr9_+_137001185 | 0.10 |
ENST00000358625.3 |
WDR5 |
WD repeat domain 5 |
| chr12_+_57623477 | 0.10 |
ENST00000557487.1 ENST00000555634.1 ENST00000556689.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr18_-_33077556 | 0.09 |
ENST00000589273.1 ENST00000586489.1 |
INO80C |
INO80 complex subunit C |
| chr9_+_111624577 | 0.09 |
ENST00000333999.3 |
ACTL7A |
actin-like 7A |
| chrX_+_149861836 | 0.09 |
ENST00000542156.1 ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1 |
myotubularin related protein 1 |
| chr1_+_110163709 | 0.09 |
ENST00000369840.2 ENST00000527846.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr3_-_183967296 | 0.09 |
ENST00000455059.1 ENST00000445626.2 |
ALG3 |
ALG3, alpha-1,3- mannosyltransferase |
| chr12_+_57623869 | 0.09 |
ENST00000414700.3 ENST00000557703.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr17_-_5342380 | 0.09 |
ENST00000225698.4 |
C1QBP |
complement component 1, q subcomponent binding protein |
| chr2_+_198365095 | 0.09 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr7_-_103629963 | 0.09 |
ENST00000428762.1 ENST00000343529.5 ENST00000424685.2 |
RELN |
reelin |
| chr1_-_227922934 | 0.09 |
ENST00000366758.3 ENST00000438896.2 |
JMJD4 |
jumonji domain containing 4 |
| chr6_-_80657292 | 0.09 |
ENST00000369816.4 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
| chr14_-_91526922 | 0.09 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr11_+_125774362 | 0.09 |
ENST00000530414.1 ENST00000530129.2 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr7_-_142099977 | 0.09 |
ENST00000390359.3 |
TRBV7-8 |
T cell receptor beta variable 7-8 |
| chr4_-_103266219 | 0.09 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr6_-_114292449 | 0.09 |
ENST00000519065.1 |
HDAC2 |
histone deacetylase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.4 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.5 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.4 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.1 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.0 | GO:1900368 | transcription, RNA-templated(GO:0001172) regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.0 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.0 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.3 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.0 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |