Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for CREB5_CREM_JUNB
Z-value: 0.06
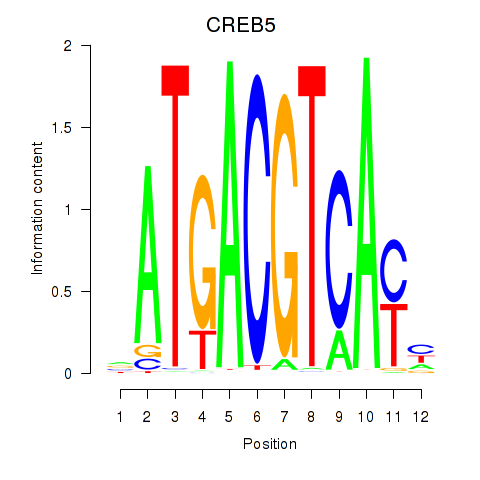
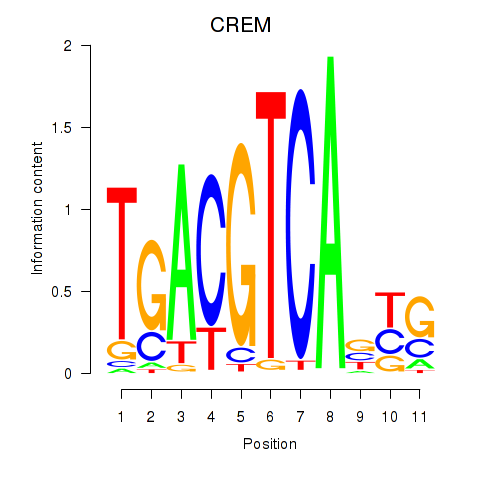
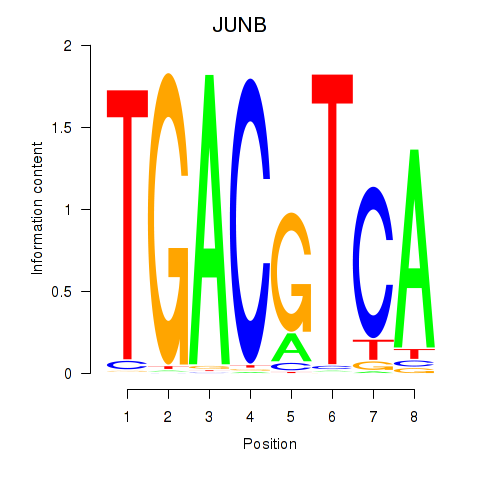
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.12 | CREB5 |
|
CREM
|
ENSG00000095794.15 | CREM |
|
JUNB
|
ENSG00000171223.4 | JUNB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUNB | hg19_v2_chr19_+_12902289_12902310 | 0.90 | 2.1e-03 | Click! |
| CREM | hg19_v2_chr10_+_35456444_35456498 | 0.61 | 1.1e-01 | Click! |
| CREB5 | hg19_v2_chr7_+_28452130_28452154 | 0.35 | 4.0e-01 | Click! |
Activity profile of CREB5_CREM_JUNB motif
Sorted Z-values of CREB5_CREM_JUNB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB5_CREM_JUNB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33146510 | 2.09 |
ENST00000397709.1 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
| chr6_+_139456226 | 1.15 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr1_+_203274639 | 1.11 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr6_+_89791507 | 0.98 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr7_+_30174426 | 0.92 |
ENST00000324453.8 |
C7orf41 |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_-_175964366 | 0.80 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr1_+_70820451 | 0.73 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr7_-_129592700 | 0.67 |
ENST00000472396.1 ENST00000355621.3 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr11_-_72496976 | 0.58 |
ENST00000539138.1 ENST00000542989.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr12_+_10365404 | 0.58 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr12_-_6772303 | 0.55 |
ENST00000396807.4 ENST00000446105.2 ENST00000341550.4 |
ING4 |
inhibitor of growth family, member 4 |
| chr16_+_29911666 | 0.55 |
ENST00000563177.1 ENST00000483405.1 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
| chr16_-_4588822 | 0.52 |
ENST00000564828.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr11_+_117070037 | 0.52 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr6_-_19804973 | 0.51 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr2_-_224467093 | 0.50 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr6_-_91006461 | 0.50 |
ENST00000257749.4 ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr16_-_4588762 | 0.49 |
ENST00000562334.1 ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr16_+_29911864 | 0.49 |
ENST00000308748.5 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
| chr2_+_220144052 | 0.48 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr14_+_68086515 | 0.48 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr22_+_25003606 | 0.48 |
ENST00000432867.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr16_-_4588469 | 0.46 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr7_-_129592471 | 0.46 |
ENST00000473814.2 ENST00000490974.1 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr17_+_6918064 | 0.46 |
ENST00000546760.1 ENST00000552402.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr6_-_46293378 | 0.46 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr5_-_172198190 | 0.45 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr20_+_17207665 | 0.44 |
ENST00000536609.1 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr6_+_56819773 | 0.44 |
ENST00000370750.2 |
BEND6 |
BEN domain containing 6 |
| chr14_+_102276192 | 0.43 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_+_6918093 | 0.42 |
ENST00000439424.2 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr17_+_6918354 | 0.41 |
ENST00000552775.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr6_-_91006627 | 0.41 |
ENST00000537989.1 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr3_-_99594948 | 0.41 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr6_+_136172820 | 0.41 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr1_-_221915418 | 0.39 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr13_-_52378231 | 0.38 |
ENST00000280056.2 ENST00000444610.2 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
| chr2_+_30454390 | 0.38 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr17_-_43209862 | 0.38 |
ENST00000322765.5 |
PLCD3 |
phospholipase C, delta 3 |
| chr17_+_41177220 | 0.37 |
ENST00000587250.2 ENST00000544533.1 |
RND2 |
Rho family GTPase 2 |
| chr12_-_4754339 | 0.36 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr16_-_79634595 | 0.36 |
ENST00000326043.4 ENST00000393350.1 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr19_+_8455077 | 0.35 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr3_+_139654018 | 0.35 |
ENST00000458420.3 |
CLSTN2 |
calsyntenin 2 |
| chr1_+_85527987 | 0.35 |
ENST00000326813.8 ENST00000294664.6 ENST00000528899.1 |
WDR63 |
WD repeat domain 63 |
| chr7_+_142031986 | 0.34 |
ENST00000547918.2 |
TRBV7-1 |
T cell receptor beta variable 7-1 (non-functional) |
| chr11_-_94965667 | 0.34 |
ENST00000542176.1 ENST00000278499.2 |
SESN3 |
sestrin 3 |
| chr3_-_21792838 | 0.33 |
ENST00000281523.2 |
ZNF385D |
zinc finger protein 385D |
| chr6_-_121655593 | 0.33 |
ENST00000398212.2 |
TBC1D32 |
TBC1 domain family, member 32 |
| chr8_+_26240414 | 0.33 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr12_-_92539614 | 0.32 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr19_+_8455200 | 0.32 |
ENST00000601897.1 ENST00000594216.1 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr11_-_111782484 | 0.31 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr10_+_35484793 | 0.31 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr17_-_4643114 | 0.31 |
ENST00000293778.6 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
| chr5_+_32174483 | 0.31 |
ENST00000606994.1 |
CTD-2186M15.3 |
CTD-2186M15.3 |
| chr10_-_64576105 | 0.31 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr17_-_7835228 | 0.30 |
ENST00000303731.4 ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1 |
trafficking protein particle complex 1 |
| chr14_+_102276132 | 0.30 |
ENST00000350249.3 ENST00000557621.1 ENST00000556946.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr19_+_35849723 | 0.30 |
ENST00000594310.1 |
FFAR3 |
free fatty acid receptor 3 |
| chr20_+_17207636 | 0.30 |
ENST00000262545.2 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr11_-_111782696 | 0.30 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr3_+_8543533 | 0.29 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr9_+_116298778 | 0.29 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr11_-_13517565 | 0.29 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr3_-_99595037 | 0.29 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr19_+_35849362 | 0.29 |
ENST00000327809.4 |
FFAR3 |
free fatty acid receptor 3 |
| chr10_+_35484053 | 0.29 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr17_+_58677539 | 0.29 |
ENST00000305921.3 |
PPM1D |
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr20_-_44420507 | 0.28 |
ENST00000243938.4 |
WFDC3 |
WAP four-disulfide core domain 3 |
| chr3_+_8543393 | 0.28 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr19_-_52227221 | 0.28 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr20_+_58179582 | 0.27 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr11_-_82782861 | 0.27 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr16_+_29973351 | 0.27 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr19_+_35861831 | 0.26 |
ENST00000454971.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr3_+_141457030 | 0.26 |
ENST00000273480.3 |
RNF7 |
ring finger protein 7 |
| chr2_+_220143989 | 0.26 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_+_37571717 | 0.26 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr2_+_37571845 | 0.25 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr1_+_26496362 | 0.25 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr3_+_159570722 | 0.25 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr19_+_35862192 | 0.25 |
ENST00000597214.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr19_+_16187085 | 0.25 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr15_+_75287861 | 0.24 |
ENST00000425597.3 ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5 |
secretory carrier membrane protein 5 |
| chr6_-_106773491 | 0.24 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr14_+_88852059 | 0.24 |
ENST00000045347.7 |
SPATA7 |
spermatogenesis associated 7 |
| chr15_-_66790146 | 0.24 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr3_+_141457105 | 0.23 |
ENST00000480908.1 ENST00000393000.3 |
RNF7 |
ring finger protein 7 |
| chr11_-_6633799 | 0.23 |
ENST00000299424.4 |
TAF10 |
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr12_-_5352315 | 0.23 |
ENST00000536518.1 |
RP11-319E16.1 |
RP11-319E16.1 |
| chr6_+_37225540 | 0.23 |
ENST00000373491.3 |
TBC1D22B |
TBC1 domain family, member 22B |
| chr17_-_40540484 | 0.23 |
ENST00000588969.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr5_-_133706695 | 0.23 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr7_-_6388537 | 0.23 |
ENST00000313324.4 ENST00000530143.1 |
FAM220A |
family with sequence similarity 220, member A |
| chr3_+_150126101 | 0.22 |
ENST00000361875.3 ENST00000361136.2 |
TSC22D2 |
TSC22 domain family, member 2 |
| chr6_-_26285737 | 0.22 |
ENST00000377727.1 ENST00000289352.1 |
HIST1H4H |
histone cluster 1, H4h |
| chr3_+_158991025 | 0.22 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr8_+_38243821 | 0.22 |
ENST00000519476.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr9_-_99381660 | 0.22 |
ENST00000375240.3 ENST00000463569.1 |
CDC14B |
cell division cycle 14B |
| chr12_+_100660909 | 0.22 |
ENST00000549687.1 |
SCYL2 |
SCY1-like 2 (S. cerevisiae) |
| chr6_-_106773610 | 0.22 |
ENST00000369076.3 ENST00000369070.1 |
ATG5 |
autophagy related 5 |
| chr5_-_178054105 | 0.21 |
ENST00000316308.4 |
CLK4 |
CDC-like kinase 4 |
| chr9_-_99382065 | 0.21 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr17_+_43209967 | 0.21 |
ENST00000431281.1 ENST00000591859.1 |
ACBD4 |
acyl-CoA binding domain containing 4 |
| chr6_-_34664612 | 0.21 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr16_+_56225248 | 0.21 |
ENST00000262493.6 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr19_+_6135646 | 0.21 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chr19_+_36024310 | 0.21 |
ENST00000222286.4 |
GAPDHS |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr9_+_34989638 | 0.21 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr15_+_76629064 | 0.21 |
ENST00000290759.4 |
ISL2 |
ISL LIM homeobox 2 |
| chr1_-_47184745 | 0.20 |
ENST00000544071.1 |
EFCAB14 |
EF-hand calcium binding domain 14 |
| chr9_+_17579084 | 0.20 |
ENST00000380607.4 |
SH3GL2 |
SH3-domain GRB2-like 2 |
| chr6_-_106773291 | 0.19 |
ENST00000343245.3 |
ATG5 |
autophagy related 5 |
| chr5_+_68513557 | 0.19 |
ENST00000256441.4 |
MRPS36 |
mitochondrial ribosomal protein S36 |
| chr17_-_7155802 | 0.19 |
ENST00000572043.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr17_+_4046462 | 0.19 |
ENST00000577075.2 ENST00000575251.1 ENST00000301391.3 |
CYB5D2 |
cytochrome b5 domain containing 2 |
| chr2_-_98280383 | 0.19 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr14_+_60715928 | 0.19 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr9_-_73029540 | 0.19 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr17_-_40540377 | 0.19 |
ENST00000404395.3 ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr17_-_74236382 | 0.19 |
ENST00000592271.1 ENST00000319945.6 ENST00000269391.6 |
RNF157 |
ring finger protein 157 |
| chr10_-_99447024 | 0.19 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr10_+_124134201 | 0.18 |
ENST00000368990.3 ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr9_-_131644202 | 0.18 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr3_+_8543561 | 0.18 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr9_+_34990219 | 0.18 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr2_-_28113965 | 0.18 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chrX_+_155110956 | 0.18 |
ENST00000286448.6 ENST00000262640.6 ENST00000460621.1 |
VAMP7 |
vesicle-associated membrane protein 7 |
| chr14_+_75746340 | 0.17 |
ENST00000555686.1 ENST00000555672.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr12_-_50790267 | 0.17 |
ENST00000327337.5 ENST00000543111.1 |
FAM186A |
family with sequence similarity 186, member A |
| chr4_+_668348 | 0.17 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr14_+_75746781 | 0.17 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr20_-_34025999 | 0.17 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr3_-_9811595 | 0.17 |
ENST00000256460.3 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
| chr19_-_59066452 | 0.17 |
ENST00000312547.2 |
CHMP2A |
charged multivesicular body protein 2A |
| chr12_+_54378923 | 0.17 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr1_-_150602035 | 0.17 |
ENST00000503241.1 ENST00000369016.4 ENST00000339643.5 ENST00000271690.8 ENST00000356527.5 ENST00000362052.7 ENST00000503345.1 ENST00000369014.5 ENST00000369009.3 |
ENSA |
endosulfine alpha |
| chr10_+_22634384 | 0.17 |
ENST00000376624.3 ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6 |
sperm associated antigen 6 |
| chr3_+_184053703 | 0.17 |
ENST00000450976.1 ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A |
family with sequence similarity 131, member A |
| chrX_+_110339439 | 0.16 |
ENST00000372010.1 ENST00000519681.1 ENST00000372007.5 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr19_+_16186903 | 0.16 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chr11_-_8832182 | 0.16 |
ENST00000527510.1 ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5 |
suppression of tumorigenicity 5 |
| chr19_-_50528584 | 0.16 |
ENST00000594092.1 ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3 |
vaccinia related kinase 3 |
| chr1_-_76398077 | 0.16 |
ENST00000284142.6 |
ASB17 |
ankyrin repeat and SOCS box containing 17 |
| chr11_+_369804 | 0.16 |
ENST00000329962.6 |
B4GALNT4 |
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr19_-_59066327 | 0.16 |
ENST00000596708.1 ENST00000601220.1 ENST00000597848.1 |
CHMP2A |
charged multivesicular body protein 2A |
| chr7_-_45128472 | 0.16 |
ENST00000490531.2 |
NACAD |
NAC alpha domain containing |
| chr4_-_668108 | 0.15 |
ENST00000304312.4 |
ATP5I |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr22_-_36357671 | 0.15 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_-_36831156 | 0.15 |
ENST00000325814.5 |
C17orf96 |
chromosome 17 open reading frame 96 |
| chr14_-_50053081 | 0.15 |
ENST00000396020.3 ENST00000245458.6 |
RPS29 |
ribosomal protein S29 |
| chr7_-_19813192 | 0.15 |
ENST00000422233.1 ENST00000433641.1 |
TMEM196 |
transmembrane protein 196 |
| chr20_-_39317868 | 0.15 |
ENST00000373313.2 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr12_+_131438443 | 0.15 |
ENST00000261654.5 |
GPR133 |
G protein-coupled receptor 133 |
| chr6_-_34855773 | 0.15 |
ENST00000420584.2 ENST00000361288.4 |
TAF11 |
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr3_+_42190714 | 0.14 |
ENST00000449246.1 |
TRAK1 |
trafficking protein, kinesin binding 1 |
| chr9_-_125027079 | 0.14 |
ENST00000417201.3 |
RBM18 |
RNA binding motif protein 18 |
| chr17_+_41476327 | 0.14 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr8_-_41522779 | 0.14 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr6_-_37225391 | 0.14 |
ENST00000356757.2 |
TMEM217 |
transmembrane protein 217 |
| chrX_+_12993202 | 0.14 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr6_+_56819895 | 0.14 |
ENST00000370748.3 |
BEND6 |
BEN domain containing 6 |
| chr19_+_13056663 | 0.14 |
ENST00000541222.1 ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
| chr11_-_2193025 | 0.14 |
ENST00000333684.5 ENST00000381178.1 ENST00000381175.1 ENST00000352909.3 |
TH |
tyrosine hydroxylase |
| chr6_-_37225367 | 0.14 |
ENST00000336655.2 |
TMEM217 |
transmembrane protein 217 |
| chr5_-_147286065 | 0.14 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr17_+_55333876 | 0.14 |
ENST00000284073.2 |
MSI2 |
musashi RNA-binding protein 2 |
| chr9_-_131644306 | 0.14 |
ENST00000302586.3 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr11_-_105948040 | 0.14 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr20_+_43211149 | 0.14 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr22_+_26879817 | 0.14 |
ENST00000215917.7 |
SRRD |
SRR1 domain containing |
| chr11_-_105948129 | 0.14 |
ENST00000526793.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr19_-_18392422 | 0.14 |
ENST00000252818.3 |
JUND |
jun D proto-oncogene |
| chr8_+_123793633 | 0.14 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr11_+_18344106 | 0.14 |
ENST00000534641.1 ENST00000525831.1 ENST00000265963.4 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr7_-_142247606 | 0.13 |
ENST00000390361.3 |
TRBV7-3 |
T cell receptor beta variable 7-3 |
| chr19_+_18043810 | 0.13 |
ENST00000445755.2 |
CCDC124 |
coiled-coil domain containing 124 |
| chr12_+_12938541 | 0.13 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr14_+_88851874 | 0.13 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr13_+_49822041 | 0.13 |
ENST00000538056.1 ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
| chr1_+_25071848 | 0.13 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr1_+_156030937 | 0.13 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr5_+_170736243 | 0.13 |
ENST00000296921.5 |
TLX3 |
T-cell leukemia homeobox 3 |
| chr5_-_95297534 | 0.13 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr8_-_21966893 | 0.13 |
ENST00000522405.1 ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr17_-_56621665 | 0.13 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr1_+_8378140 | 0.13 |
ENST00000377479.2 |
SLC45A1 |
solute carrier family 45, member 1 |
| chr5_+_68513622 | 0.13 |
ENST00000512880.1 ENST00000602380.1 |
MRPS36 |
mitochondrial ribosomal protein S36 |
| chr1_-_145382362 | 0.13 |
ENST00000419817.1 ENST00000421937.3 ENST00000433081.2 |
RP11-458D21.1 |
RP11-458D21.1 |
| chr2_+_74757050 | 0.13 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr11_-_5531215 | 0.13 |
ENST00000311659.4 |
UBQLN3 |
ubiquilin 3 |
| chr1_+_29563011 | 0.13 |
ENST00000345512.3 ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU |
protein tyrosine phosphatase, receptor type, U |
| chr5_-_78809950 | 0.13 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr4_-_142054590 | 0.12 |
ENST00000306799.3 |
RNF150 |
ring finger protein 150 |
| chr15_-_83240507 | 0.12 |
ENST00000564522.1 ENST00000398592.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr16_-_86588627 | 0.12 |
ENST00000565482.1 ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD |
methenyltetrahydrofolate synthetase domain containing |
| chr15_+_44084040 | 0.12 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr6_-_159065741 | 0.12 |
ENST00000367085.3 ENST00000367089.3 |
DYNLT1 |
dynein, light chain, Tctex-type 1 |
| chr1_-_209979465 | 0.12 |
ENST00000542854.1 |
IRF6 |
interferon regulatory factor 6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 0.5 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.6 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.5 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 1.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.6 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.9 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0043605 | allantoin metabolic process(GO:0000255) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0060067 | cervix development(GO:0060067) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 1.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.5 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 2.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |