Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for DBP
Z-value: 1.19
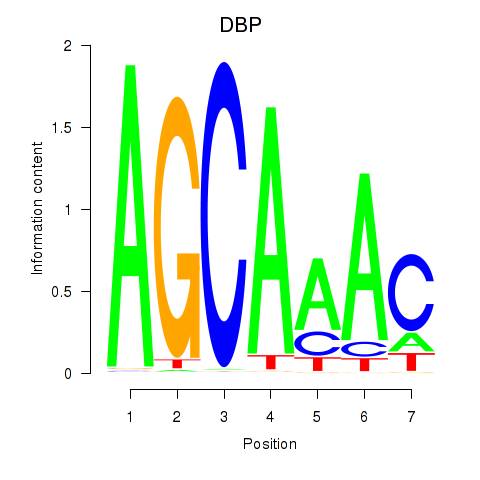
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | DBP |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49137762_49137783, hg19_v2_chr19_-_49140609_49140643 | -0.56 | 1.5e-01 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 1.70 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr1_+_114522049 | 1.22 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr6_-_117747015 | 1.18 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr3_+_141105705 | 1.13 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr9_-_95186739 | 1.00 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr2_-_190044480 | 0.95 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr8_+_104384616 | 0.90 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr12_-_56236734 | 0.90 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr8_-_120685608 | 0.88 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_111091948 | 0.87 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr13_-_67802549 | 0.84 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr10_-_79397202 | 0.83 |
ENST00000372437.1 ENST00000372408.2 ENST00000372403.4 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79397479 | 0.80 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79397316 | 0.79 |
ENST00000372421.5 ENST00000457953.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_56236690 | 0.74 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr4_-_57547454 | 0.74 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr2_+_210444748 | 0.71 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr9_-_89562104 | 0.65 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr3_-_114343768 | 0.64 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr10_-_79397391 | 0.63 |
ENST00000286628.8 ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_+_210444142 | 0.61 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr5_-_124080203 | 0.60 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr1_+_223101757 | 0.59 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr13_+_93879085 | 0.59 |
ENST00000377047.4 |
GPC6 |
glypican 6 |
| chr4_+_55095264 | 0.58 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chrX_+_149531524 | 0.53 |
ENST00000370401.2 |
MAMLD1 |
mastermind-like domain containing 1 |
| chr3_+_141106458 | 0.52 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr18_-_25616519 | 0.50 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr6_-_56819385 | 0.50 |
ENST00000370754.5 ENST00000449297.2 |
DST |
dystonin |
| chr3_+_141043050 | 0.49 |
ENST00000509842.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr16_+_86612112 | 0.48 |
ENST00000320241.3 |
FOXL1 |
forkhead box L1 |
| chr2_+_210517895 | 0.45 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr5_-_41510656 | 0.45 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_41510725 | 0.44 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_+_61547405 | 0.41 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr9_+_118916082 | 0.40 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr8_+_16884740 | 0.39 |
ENST00000318063.5 |
MICU3 |
mitochondrial calcium uptake family, member 3 |
| chr3_+_69812701 | 0.38 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr1_-_85870177 | 0.38 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr5_-_158526693 | 0.36 |
ENST00000380654.4 |
EBF1 |
early B-cell factor 1 |
| chr13_-_114843416 | 0.35 |
ENST00000389544.4 |
RASA3 |
RAS p21 protein activator 3 |
| chr3_+_69812877 | 0.35 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr5_-_158526756 | 0.33 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr6_-_31514516 | 0.33 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr6_-_31514333 | 0.33 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr17_-_8263538 | 0.33 |
ENST00000535173.1 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr8_+_77593474 | 0.33 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr5_+_32710736 | 0.32 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr4_-_157892167 | 0.32 |
ENST00000541126.1 |
PDGFC |
platelet derived growth factor C |
| chr5_+_140710061 | 0.31 |
ENST00000517417.1 ENST00000378105.3 |
PCDHGA1 |
protocadherin gamma subfamily A, 1 |
| chr4_-_157892055 | 0.30 |
ENST00000422544.2 |
PDGFC |
platelet derived growth factor C |
| chr3_-_145878954 | 0.28 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr1_+_104293028 | 0.28 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr7_-_150946015 | 0.27 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr4_+_113970772 | 0.27 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr14_-_37051798 | 0.27 |
ENST00000258829.5 |
NKX2-8 |
NK2 homeobox 8 |
| chr1_-_227505289 | 0.27 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr4_+_106473768 | 0.26 |
ENST00000265154.2 ENST00000420470.2 |
ARHGEF38 |
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr1_+_104159999 | 0.26 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr14_+_29234870 | 0.25 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr4_-_157892498 | 0.25 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr20_+_31595406 | 0.25 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr10_-_64576105 | 0.25 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr13_-_41593425 | 0.23 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_211433275 | 0.23 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr4_-_100242549 | 0.23 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr2_+_95963052 | 0.22 |
ENST00000295225.5 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
| chr12_-_92536433 | 0.22 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr9_+_2158443 | 0.22 |
ENST00000302401.3 ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_104178946 | 0.22 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr4_+_114214125 | 0.22 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chr3_+_152017360 | 0.22 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr4_+_160188889 | 0.21 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_114343039 | 0.21 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_179233376 | 0.21 |
ENST00000376929.3 ENST00000514093.1 |
SQSTM1 |
sequestosome 1 |
| chr6_+_10528560 | 0.21 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr12_+_56511943 | 0.20 |
ENST00000257940.2 ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10 ESYT1 |
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr6_+_3118926 | 0.20 |
ENST00000380379.5 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
| chr15_-_63450192 | 0.20 |
ENST00000411926.1 |
RPS27L |
ribosomal protein S27-like |
| chr19_+_11455900 | 0.20 |
ENST00000588790.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr20_+_11898507 | 0.20 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr5_+_72143988 | 0.19 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr1_+_101361782 | 0.19 |
ENST00000357650.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr11_-_66445219 | 0.19 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr9_+_2158485 | 0.19 |
ENST00000417599.1 ENST00000382185.1 ENST00000382183.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr22_-_31742218 | 0.19 |
ENST00000266269.5 ENST00000405309.3 ENST00000351933.4 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_+_51026743 | 0.18 |
ENST00000358385.6 ENST00000357032.3 ENST00000354525.4 |
ATL1 |
atlastin GTPase 1 |
| chr11_+_35684288 | 0.18 |
ENST00000299413.5 |
TRIM44 |
tripartite motif containing 44 |
| chr1_+_153700518 | 0.18 |
ENST00000318967.2 ENST00000456435.1 ENST00000435409.2 |
INTS3 |
integrator complex subunit 3 |
| chr10_-_31288398 | 0.18 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr21_+_17791838 | 0.17 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr6_-_24936170 | 0.17 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr1_+_84609944 | 0.17 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_211036051 | 0.17 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr3_+_152017181 | 0.17 |
ENST00000498502.1 ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr1_+_61548225 | 0.17 |
ENST00000371187.3 |
NFIA |
nuclear factor I/A |
| chr5_-_142780280 | 0.16 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr16_-_18468926 | 0.16 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr2_-_216257849 | 0.16 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr10_+_35484053 | 0.16 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr12_+_7169887 | 0.16 |
ENST00000542978.1 |
C1S |
complement component 1, s subcomponent |
| chr21_+_17791648 | 0.16 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr7_+_13141097 | 0.15 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr5_-_58295712 | 0.15 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr2_-_164592497 | 0.15 |
ENST00000333129.3 ENST00000409634.1 |
FIGN |
fidgetin |
| chr5_-_137674000 | 0.15 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr4_+_88754113 | 0.14 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr15_+_67418047 | 0.14 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr6_+_33168637 | 0.14 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr5_+_140588269 | 0.14 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr9_-_13165457 | 0.14 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr10_+_21823243 | 0.14 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr7_-_121784285 | 0.14 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr14_+_58765103 | 0.14 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr5_+_180682720 | 0.14 |
ENST00000599439.1 |
AC008443.1 |
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr13_+_44947941 | 0.13 |
ENST00000379179.3 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
| chr6_+_33168597 | 0.13 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_42580738 | 0.13 |
ENST00000585614.1 ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8 |
G patch domain containing 8 |
| chr17_+_2240775 | 0.13 |
ENST00000268989.3 ENST00000426855.2 |
SGSM2 |
small G protein signaling modulator 2 |
| chr3_+_63953415 | 0.12 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr10_-_14050522 | 0.12 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr16_+_89724188 | 0.12 |
ENST00000301031.4 ENST00000566204.1 ENST00000579310.1 |
SPATA33 |
spermatogenesis associated 33 |
| chr3_-_47950745 | 0.12 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr17_+_79953310 | 0.12 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr8_-_29120580 | 0.12 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr17_+_2240916 | 0.12 |
ENST00000574563.1 |
SGSM2 |
small G protein signaling modulator 2 |
| chr14_+_24590560 | 0.12 |
ENST00000558325.1 |
RP11-468E2.6 |
RP11-468E2.6 |
| chrX_+_28605516 | 0.11 |
ENST00000378993.1 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
| chr14_+_29236269 | 0.11 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr10_+_47894572 | 0.11 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr19_+_41856816 | 0.11 |
ENST00000539627.1 |
TMEM91 |
transmembrane protein 91 |
| chrX_-_24045303 | 0.11 |
ENST00000328046.8 |
KLHL15 |
kelch-like family member 15 |
| chr4_+_88754069 | 0.11 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr15_+_71228826 | 0.11 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr9_+_131037623 | 0.11 |
ENST00000495313.1 ENST00000372898.2 |
SWI5 |
SWI5 recombination repair homolog (yeast) |
| chr12_+_18891045 | 0.10 |
ENST00000317658.3 |
CAPZA3 |
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr1_-_114414316 | 0.10 |
ENST00000528414.1 ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr9_+_35829208 | 0.10 |
ENST00000439587.2 ENST00000377991.4 |
TMEM8B |
transmembrane protein 8B |
| chr16_-_75282088 | 0.10 |
ENST00000542031.2 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
| chr15_-_27018175 | 0.09 |
ENST00000311550.5 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr4_+_71587669 | 0.09 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr19_-_39881777 | 0.09 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr8_+_68864330 | 0.09 |
ENST00000288368.4 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr6_+_31514622 | 0.09 |
ENST00000376146.4 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr3_-_88108212 | 0.09 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr1_+_206557366 | 0.09 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr10_-_104178857 | 0.08 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr3_-_71353892 | 0.08 |
ENST00000484350.1 |
FOXP1 |
forkhead box P1 |
| chr12_+_56137064 | 0.08 |
ENST00000257868.5 ENST00000546799.1 |
GDF11 |
growth differentiation factor 11 |
| chr9_+_72658490 | 0.08 |
ENST00000377182.4 |
MAMDC2 |
MAM domain containing 2 |
| chrX_+_129473916 | 0.08 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr16_+_30960375 | 0.08 |
ENST00000318663.4 ENST00000566237.1 ENST00000562699.1 |
ORAI3 |
ORAI calcium release-activated calcium modulator 3 |
| chr8_+_26247878 | 0.08 |
ENST00000518611.1 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr9_+_82187630 | 0.08 |
ENST00000265284.6 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr11_-_115630900 | 0.08 |
ENST00000537070.1 ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900 |
long intergenic non-protein coding RNA 900 |
| chr4_-_140098339 | 0.08 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr1_+_61547894 | 0.08 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr14_+_45366472 | 0.08 |
ENST00000325192.3 |
C14orf28 |
chromosome 14 open reading frame 28 |
| chr5_+_161495038 | 0.08 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_-_88427568 | 0.08 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr15_+_38544476 | 0.08 |
ENST00000299084.4 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
| chr10_-_65028817 | 0.08 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chrX_+_10031499 | 0.08 |
ENST00000454666.1 |
WWC3 |
WWC family member 3 |
| chr10_-_29811456 | 0.08 |
ENST00000535393.1 |
SVIL |
supervillin |
| chr1_+_154229547 | 0.08 |
ENST00000428595.1 |
UBAP2L |
ubiquitin associated protein 2-like |
| chr9_+_82186872 | 0.07 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_+_39881951 | 0.07 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr3_+_153202284 | 0.07 |
ENST00000446603.2 |
C3orf79 |
chromosome 3 open reading frame 79 |
| chr19_-_42806919 | 0.07 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr9_+_131038425 | 0.07 |
ENST00000320188.5 ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5 |
SWI5 recombination repair homolog (yeast) |
| chr11_-_94706705 | 0.07 |
ENST00000279839.6 |
CWC15 |
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr8_+_85095769 | 0.07 |
ENST00000518566.1 |
RALYL |
RALY RNA binding protein-like |
| chr16_+_25228242 | 0.07 |
ENST00000219660.5 |
AQP8 |
aquaporin 8 |
| chr22_+_31742875 | 0.07 |
ENST00000504184.2 |
AC005003.1 |
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr16_+_30675654 | 0.07 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr3_+_185300391 | 0.07 |
ENST00000545472.1 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr1_-_196577489 | 0.07 |
ENST00000609185.1 ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2 |
potassium channel, subfamily T, member 2 |
| chr4_+_71588372 | 0.07 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr19_-_39881669 | 0.07 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_+_13135386 | 0.07 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_+_67022633 | 0.07 |
ENST00000398354.1 ENST00000326686.5 |
CES4A |
carboxylesterase 4A |
| chr16_+_67022476 | 0.07 |
ENST00000540947.2 |
CES4A |
carboxylesterase 4A |
| chr16_+_67022582 | 0.07 |
ENST00000541479.1 ENST00000338718.4 |
CES4A |
carboxylesterase 4A |
| chr3_-_88108192 | 0.07 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr11_+_102980251 | 0.06 |
ENST00000334267.7 ENST00000398093.3 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr3_+_88108381 | 0.06 |
ENST00000473136.1 |
RP11-159G9.5 |
Uncharacterized protein |
| chr7_+_99613212 | 0.06 |
ENST00000426572.1 ENST00000535170.1 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr17_+_4613918 | 0.06 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chr9_+_82187487 | 0.06 |
ENST00000435650.1 ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr12_-_89918522 | 0.06 |
ENST00000529983.2 |
GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr3_+_140396881 | 0.06 |
ENST00000286349.3 |
TRIM42 |
tripartite motif containing 42 |
| chr3_+_109128837 | 0.06 |
ENST00000497996.1 |
RP11-702L6.4 |
RP11-702L6.4 |
| chr1_+_203734296 | 0.06 |
ENST00000442561.2 ENST00000367217.5 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
| chr19_+_10765699 | 0.06 |
ENST00000590009.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr10_-_65028938 | 0.06 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr4_-_74088800 | 0.06 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr2_-_21266935 | 0.06 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr20_-_18447667 | 0.06 |
ENST00000262547.5 ENST00000329494.5 ENST00000357236.4 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
| chr5_+_129083772 | 0.05 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 3.1 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 1.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 1.6 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.0 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.0 | 0.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |