Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for DBX2_HLX
Z-value: 0.08
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
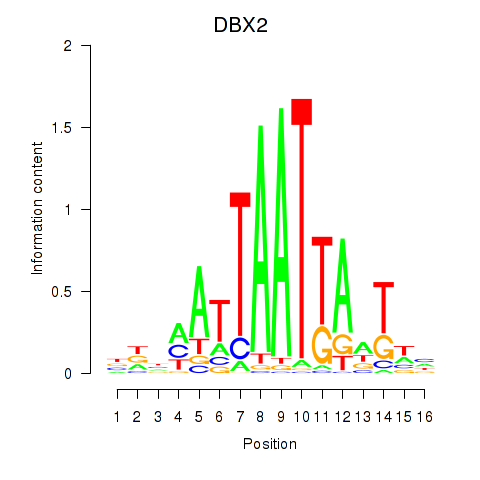
DBX2
|
ENSG00000185610.6 | DBX2 |
|
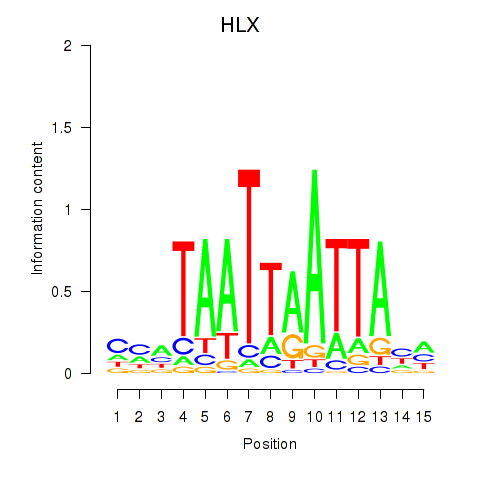
HLX
|
ENSG00000136630.11 | HLX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg19_v2_chr1_+_221051699_221051699 | 0.22 | 6.0e-01 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_112564797 | 0.38 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr9_-_95166841 | 0.37 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr9_-_95166884 | 0.34 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr5_+_55149150 | 0.32 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr12_+_26348429 | 0.26 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr12_+_26348246 | 0.24 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr1_+_81771806 | 0.23 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr17_+_1674982 | 0.21 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_-_93115445 | 0.20 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chrX_-_15332665 | 0.20 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr7_-_140482926 | 0.19 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr8_+_104892639 | 0.18 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_+_67498538 | 0.17 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr21_-_31852663 | 0.16 |
ENST00000390689.2 |
KRTAP19-1 |
keratin associated protein 19-1 |
| chr10_+_70847852 | 0.15 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr11_-_63376013 | 0.15 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr8_+_97597148 | 0.15 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr9_-_95244781 | 0.14 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr3_-_58613323 | 0.14 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr12_-_10573149 | 0.14 |
ENST00000381904.2 ENST00000381903.2 ENST00000396439.2 |
KLRC3 |
killer cell lectin-like receptor subfamily C, member 3 |
| chr3_-_114790179 | 0.14 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_+_5372738 | 0.14 |
ENST00000380219.1 |
OR51B6 |
olfactory receptor, family 51, subfamily B, member 6 |
| chr14_-_45252031 | 0.14 |
ENST00000556405.1 |
RP11-398E10.1 |
RP11-398E10.1 |
| chr4_+_71108300 | 0.13 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr15_+_67418047 | 0.13 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr10_+_69865866 | 0.13 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr5_-_20575959 | 0.13 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr10_-_27529486 | 0.13 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr8_-_116504448 | 0.12 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr15_+_94899183 | 0.12 |
ENST00000557742.1 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
| chr2_+_161993465 | 0.12 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_91152303 | 0.12 |
ENST00000371804.3 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 |
| chr1_+_196788887 | 0.11 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr2_-_225811747 | 0.11 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr4_+_70916119 | 0.11 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr11_-_13517565 | 0.11 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr10_-_29084886 | 0.10 |
ENST00000608061.1 ENST00000443246.2 ENST00000446012.1 |
LINC00837 |
long intergenic non-protein coding RNA 837 |
| chr10_-_4285923 | 0.10 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr15_+_22382382 | 0.10 |
ENST00000328795.4 |
OR4N4 |
olfactory receptor, family 4, subfamily N, member 4 |
| chr11_-_26593779 | 0.10 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr2_+_161993412 | 0.09 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr11_-_26593677 | 0.09 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chrX_+_36254051 | 0.09 |
ENST00000378657.4 |
CXorf30 |
chromosome X open reading frame 30 |
| chr7_-_14942944 | 0.09 |
ENST00000403951.2 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr7_+_142498725 | 0.09 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr10_-_28571015 | 0.09 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr19_+_42212526 | 0.08 |
ENST00000598976.1 ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA CEACAM5 |
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chrX_+_55744228 | 0.08 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr4_-_138453606 | 0.08 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr19_+_54466179 | 0.08 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr12_-_91505608 | 0.08 |
ENST00000266718.4 |
LUM |
lumican |
| chrX_+_55744166 | 0.08 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr3_+_138340067 | 0.08 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr18_+_6834472 | 0.08 |
ENST00000581099.1 ENST00000419673.2 ENST00000531294.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr6_+_10521574 | 0.08 |
ENST00000495262.1 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr2_+_166095898 | 0.08 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_+_36246735 | 0.08 |
ENST00000378653.3 |
CXorf30 |
chromosome X open reading frame 30 |
| chr7_+_129984630 | 0.08 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr2_-_145277569 | 0.07 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr4_-_159080806 | 0.07 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr4_-_90757364 | 0.07 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_101918153 | 0.07 |
ENST00000434758.2 ENST00000526781.1 ENST00000534360.1 |
C11orf70 |
chromosome 11 open reading frame 70 |
| chr6_+_27925019 | 0.07 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr2_+_26624775 | 0.07 |
ENST00000288710.2 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr4_-_152149033 | 0.06 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr5_+_95066823 | 0.06 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr12_+_64798095 | 0.06 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr6_-_161085291 | 0.06 |
ENST00000316300.5 |
LPA |
lipoprotein, Lp(a) |
| chr4_-_138453559 | 0.06 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr1_+_225600404 | 0.06 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr10_+_81891416 | 0.06 |
ENST00000372270.2 |
PLAC9 |
placenta-specific 9 |
| chr6_+_31021225 | 0.06 |
ENST00000565192.1 ENST00000562344.1 |
HCG22 |
HLA complex group 22 |
| chr10_-_28623368 | 0.06 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_-_110986918 | 0.05 |
ENST00000297404.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr12_+_12510352 | 0.05 |
ENST00000298571.6 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
| chr8_-_145754428 | 0.05 |
ENST00000527462.1 ENST00000313465.5 ENST00000524821.1 |
C8orf82 |
chromosome 8 open reading frame 82 |
| chr6_+_121756809 | 0.05 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr2_-_179343226 | 0.05 |
ENST00000434643.2 |
FKBP7 |
FK506 binding protein 7 |
| chr9_+_105757590 | 0.05 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr11_-_327537 | 0.05 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr9_-_21351377 | 0.05 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr2_+_103035102 | 0.05 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr8_+_11961898 | 0.05 |
ENST00000400085.3 |
ZNF705D |
zinc finger protein 705D |
| chr7_+_141811539 | 0.05 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr1_+_196912902 | 0.05 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr4_+_74347400 | 0.05 |
ENST00000226355.3 |
AFM |
afamin |
| chr4_-_163085107 | 0.05 |
ENST00000379164.4 |
FSTL5 |
follistatin-like 5 |
| chr14_-_106552755 | 0.05 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr1_-_231005310 | 0.05 |
ENST00000470540.1 |
C1orf198 |
chromosome 1 open reading frame 198 |
| chr1_-_204183071 | 0.05 |
ENST00000308302.3 |
GOLT1A |
golgi transport 1A |
| chr2_+_219472488 | 0.05 |
ENST00000450993.2 |
PLCD4 |
phospholipase C, delta 4 |
| chr11_-_2162468 | 0.05 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr6_+_134758827 | 0.05 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr1_+_221051699 | 0.05 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr8_-_101157680 | 0.04 |
ENST00000428847.2 |
FBXO43 |
F-box protein 43 |
| chr5_-_131879205 | 0.04 |
ENST00000231454.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr12_+_8309630 | 0.04 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr19_-_14992264 | 0.04 |
ENST00000327462.2 |
OR7A17 |
olfactory receptor, family 7, subfamily A, member 17 |
| chr1_-_160549235 | 0.04 |
ENST00000368054.3 ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84 |
CD84 molecule |
| chr4_+_71063641 | 0.04 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr7_-_92777606 | 0.04 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr19_-_58864848 | 0.04 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chrX_+_43515467 | 0.04 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr16_-_66584059 | 0.04 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr1_-_110283138 | 0.04 |
ENST00000256594.3 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr4_-_89744314 | 0.04 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr5_+_54455946 | 0.04 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr12_-_10588539 | 0.04 |
ENST00000381902.2 ENST00000381901.1 ENST00000539033.1 |
KLRC2 NKG2-E |
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr4_-_100356291 | 0.04 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr9_+_34652164 | 0.04 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr10_+_5135981 | 0.04 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr2_-_158300556 | 0.04 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr10_+_90660832 | 0.04 |
ENST00000371924.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr17_+_17685422 | 0.04 |
ENST00000395774.1 |
RAI1 |
retinoic acid induced 1 |
| chr4_+_119809984 | 0.04 |
ENST00000307142.4 ENST00000448416.2 ENST00000429713.2 |
SYNPO2 |
synaptopodin 2 |
| chr8_+_62200509 | 0.04 |
ENST00000519846.1 ENST00000518592.1 ENST00000325897.4 |
CLVS1 |
clavesin 1 |
| chr2_+_143635067 | 0.04 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr8_-_13134045 | 0.04 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr2_+_210517895 | 0.04 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr5_+_57787254 | 0.04 |
ENST00000502276.1 ENST00000396776.2 ENST00000511930.1 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
| chr6_-_112575687 | 0.04 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr16_-_3350614 | 0.04 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr18_-_40857493 | 0.04 |
ENST00000255224.3 |
SYT4 |
synaptotagmin IV |
| chr16_+_14802801 | 0.04 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr1_+_192127578 | 0.04 |
ENST00000367460.3 |
RGS18 |
regulator of G-protein signaling 18 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr11_+_120973375 | 0.04 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr14_+_37641012 | 0.03 |
ENST00000556667.1 |
SLC25A21-AS1 |
SLC25A21 antisense RNA 1 |
| chr1_-_157567868 | 0.03 |
ENST00000271532.1 |
FCRL4 |
Fc receptor-like 4 |
| chr1_-_48937838 | 0.03 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr7_+_130126165 | 0.03 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr20_+_15177480 | 0.03 |
ENST00000402914.1 |
MACROD2 |
MACRO domain containing 2 |
| chr18_-_7117813 | 0.03 |
ENST00000389658.3 |
LAMA1 |
laminin, alpha 1 |
| chr10_+_91061712 | 0.03 |
ENST00000371826.3 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
| chr6_-_52859046 | 0.03 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr15_-_75748115 | 0.03 |
ENST00000360439.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr10_-_52645379 | 0.03 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr1_+_202385953 | 0.03 |
ENST00000466968.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr8_-_101571964 | 0.03 |
ENST00000520552.1 ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr13_+_78315295 | 0.03 |
ENST00000351546.3 |
SLAIN1 |
SLAIN motif family, member 1 |
| chr5_-_36301984 | 0.03 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr11_-_26593649 | 0.03 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chrX_+_105192423 | 0.03 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr12_+_80750134 | 0.03 |
ENST00000546620.1 |
OTOGL |
otogelin-like |
| chr8_-_101571933 | 0.03 |
ENST00000520311.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr17_-_39254391 | 0.03 |
ENST00000333822.4 |
KRTAP4-8 |
keratin associated protein 4-8 |
| chr9_+_125133315 | 0.03 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_+_130126012 | 0.03 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr14_+_92789498 | 0.03 |
ENST00000531433.1 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr14_-_107049312 | 0.03 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr7_-_14026063 | 0.03 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr4_-_89744365 | 0.03 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr3_-_194072019 | 0.03 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr1_-_197036364 | 0.03 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr17_+_18647326 | 0.03 |
ENST00000395667.1 ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10 |
F-box and WD repeat domain containing 10 |
| chr19_-_4535233 | 0.03 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr15_+_25200074 | 0.03 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr16_-_20364122 | 0.03 |
ENST00000396138.4 ENST00000577168.1 |
UMOD |
uromodulin |
| chr1_+_234765057 | 0.03 |
ENST00000429269.1 |
LINC00184 |
long intergenic non-protein coding RNA 184 |
| chr9_+_27109392 | 0.03 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr19_-_51920952 | 0.03 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr3_-_48130707 | 0.03 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr12_-_10151773 | 0.03 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr20_-_29978286 | 0.03 |
ENST00000376315.2 |
DEFB119 |
defensin, beta 119 |
| chr8_+_107738343 | 0.03 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chrX_+_15525426 | 0.03 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr6_-_112575838 | 0.03 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr6_+_124125286 | 0.03 |
ENST00000368416.1 ENST00000368417.1 ENST00000546092.1 |
NKAIN2 |
Na+/K+ transporting ATPase interacting 2 |
| chr3_+_40518599 | 0.03 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr1_+_84630645 | 0.03 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_179343268 | 0.03 |
ENST00000424785.2 |
FKBP7 |
FK506 binding protein 7 |
| chr5_-_55412774 | 0.03 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr15_+_25200108 | 0.03 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr4_-_130692631 | 0.03 |
ENST00000500092.2 ENST00000509105.1 |
RP11-519M16.1 |
RP11-519M16.1 |
| chr9_-_93405352 | 0.03 |
ENST00000375765.3 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
| chr16_-_28937027 | 0.03 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr6_+_111408698 | 0.03 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr16_-_15474904 | 0.03 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr5_+_43602750 | 0.03 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr4_-_170533723 | 0.03 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr8_+_42396274 | 0.02 |
ENST00000438528.3 |
SMIM19 |
small integral membrane protein 19 |
| chr12_+_57623477 | 0.02 |
ENST00000557487.1 ENST00000555634.1 ENST00000556689.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chrX_-_13835147 | 0.02 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr5_+_36608422 | 0.02 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_-_51796987 | 0.02 |
ENST00000262676.5 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr3_+_57881966 | 0.02 |
ENST00000495364.1 |
SLMAP |
sarcolemma associated protein |
| chr8_+_19796381 | 0.02 |
ENST00000524029.1 ENST00000522701.1 ENST00000311322.8 |
LPL |
lipoprotein lipase |
| chr6_-_52668605 | 0.02 |
ENST00000334575.5 |
GSTA1 |
glutathione S-transferase alpha 1 |
| chr7_+_134576317 | 0.02 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr7_-_111032971 | 0.02 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_60058458 | 0.02 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_92371839 | 0.02 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr14_-_53331239 | 0.02 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr12_-_76817036 | 0.02 |
ENST00000546946.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr3_-_141747950 | 0.02 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_212965170 | 0.02 |
ENST00000532324.1 ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3 |
TatD DNase domain containing 3 |
| chr7_+_80275953 | 0.02 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr16_-_66583701 | 0.02 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr7_+_123241908 | 0.02 |
ENST00000434204.1 ENST00000437535.1 ENST00000451215.1 |
ASB15 |
ankyrin repeat and SOCS box containing 15 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.0 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |