Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for DLX4_HOXD8
Z-value: 0.30
Transcription factors associated with DLX4_HOXD8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
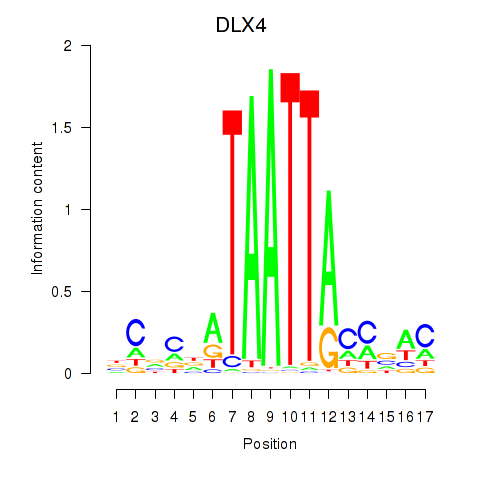
DLX4
|
ENSG00000108813.9 | DLX4 |
|
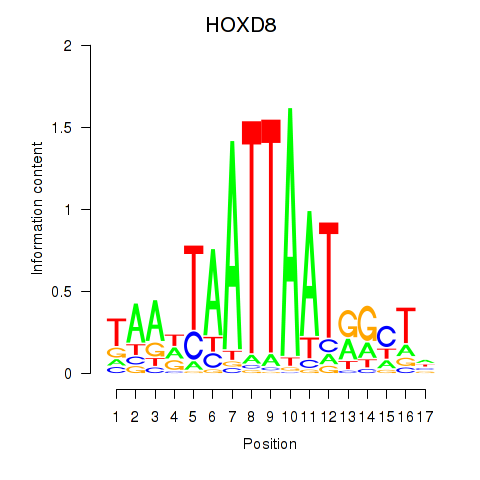
HOXD8
|
ENSG00000175879.7 | HOXD8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD8 | hg19_v2_chr2_+_176994408_176994641 | -0.61 | 1.1e-01 | Click! |
| DLX4 | hg19_v2_chr17_+_48046538_48046575 | -0.04 | 9.3e-01 | Click! |
Activity profile of DLX4_HOXD8 motif
Sorted Z-values of DLX4_HOXD8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX4_HOXD8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25801041 | 1.13 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr12_-_91573132 | 0.83 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91546926 | 0.65 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_225811747 | 0.62 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr14_-_106471723 | 0.49 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr12_-_10978957 | 0.46 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr1_-_72566613 | 0.46 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr3_+_157154578 | 0.43 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr5_-_20575959 | 0.42 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr10_-_79398250 | 0.40 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_91573316 | 0.36 |
ENST00000393155.1 |
DCN |
decorin |
| chr2_-_238323007 | 0.34 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr1_+_144989309 | 0.34 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr4_-_70626314 | 0.34 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr8_-_13134045 | 0.32 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr5_-_146781153 | 0.32 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr21_-_31744557 | 0.32 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr9_-_27005686 | 0.31 |
ENST00000380055.5 |
LRRC19 |
leucine rich repeat containing 19 |
| chr6_-_87804815 | 0.30 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr21_-_31869451 | 0.29 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr19_+_21579958 | 0.29 |
ENST00000339914.6 ENST00000599461.1 |
ZNF493 |
zinc finger protein 493 |
| chrX_-_92928557 | 0.28 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr3_-_112320749 | 0.28 |
ENST00000610103.1 |
RP11-572C15.6 |
RP11-572C15.6 |
| chr16_+_56995854 | 0.28 |
ENST00000566128.1 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr13_+_102142296 | 0.28 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_-_156787425 | 0.28 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr12_-_91573249 | 0.27 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr12_-_11214893 | 0.27 |
ENST00000533467.1 |
TAS2R46 |
taste receptor, type 2, member 46 |
| chr11_-_121986923 | 0.27 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr4_-_119759795 | 0.26 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr8_+_38585704 | 0.26 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr7_+_107224364 | 0.26 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr1_+_163039143 | 0.25 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr4_+_56815102 | 0.25 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr17_-_64216748 | 0.25 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_-_197115818 | 0.25 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr5_+_42756903 | 0.24 |
ENST00000361970.5 ENST00000388827.4 |
CCDC152 |
coiled-coil domain containing 152 |
| chr9_-_21482312 | 0.23 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr4_+_70916119 | 0.23 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr11_-_124670550 | 0.23 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr16_+_22517166 | 0.23 |
ENST00000356156.3 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr6_+_151646800 | 0.22 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr14_-_106926724 | 0.22 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr9_+_120466610 | 0.22 |
ENST00000394487.4 |
TLR4 |
toll-like receptor 4 |
| chr17_-_64225508 | 0.22 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr5_-_88119580 | 0.21 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr15_-_55657428 | 0.21 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr22_+_39353527 | 0.21 |
ENST00000249116.2 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr15_-_20170354 | 0.21 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr12_+_109826524 | 0.21 |
ENST00000431443.2 |
MYO1H |
myosin IH |
| chr6_+_153552455 | 0.21 |
ENST00000392385.2 |
AL590867.1 |
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr1_-_92371839 | 0.21 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr2_+_201450591 | 0.20 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr5_+_156696362 | 0.20 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr10_+_120116527 | 0.20 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr2_-_158345462 | 0.19 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr3_-_191000172 | 0.19 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr14_+_22580233 | 0.19 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr3_-_12587055 | 0.19 |
ENST00000564146.3 |
C3orf83 |
chromosome 3 open reading frame 83 |
| chr15_+_92006567 | 0.19 |
ENST00000554333.1 |
RP11-661P17.1 |
RP11-661P17.1 |
| chr16_+_56995762 | 0.19 |
ENST00000200676.3 ENST00000379780.2 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr20_-_8000426 | 0.19 |
ENST00000527925.1 ENST00000246024.2 |
TMX4 |
thioredoxin-related transmembrane protein 4 |
| chr4_-_74853897 | 0.18 |
ENST00000296028.3 |
PPBP |
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr10_+_32873190 | 0.18 |
ENST00000375025.4 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr14_-_92198403 | 0.18 |
ENST00000553329.1 ENST00000256343.3 |
CATSPERB |
catsper channel auxiliary subunit beta |
| chr19_-_9003586 | 0.18 |
ENST00000380951.5 |
MUC16 |
mucin 16, cell surface associated |
| chr2_+_204801471 | 0.18 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr8_+_27631903 | 0.18 |
ENST00000305188.8 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr5_+_156712372 | 0.18 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr11_-_89223883 | 0.17 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_-_100712352 | 0.17 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr12_+_51818749 | 0.17 |
ENST00000514353.3 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr1_-_27998689 | 0.17 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr6_-_117747015 | 0.17 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr20_+_56964169 | 0.16 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_+_124320195 | 0.16 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr12_-_24737089 | 0.16 |
ENST00000483544.1 |
LINC00477 |
long intergenic non-protein coding RNA 477 |
| chr5_-_147286065 | 0.16 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr16_+_24549014 | 0.16 |
ENST00000564314.1 ENST00000567686.1 |
RBBP6 |
retinoblastoma binding protein 6 |
| chr22_+_22516550 | 0.16 |
ENST00000390284.2 |
IGLV4-60 |
immunoglobulin lambda variable 4-60 |
| chr3_+_45927994 | 0.16 |
ENST00000357632.2 ENST00000395963.2 |
CCR9 |
chemokine (C-C motif) receptor 9 |
| chr13_-_38172863 | 0.16 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr1_+_240177627 | 0.16 |
ENST00000447095.1 |
FMN2 |
formin 2 |
| chr1_-_145826450 | 0.16 |
ENST00000462900.2 |
GPR89A |
G protein-coupled receptor 89A |
| chr11_-_89224139 | 0.16 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr2_+_228736321 | 0.15 |
ENST00000309931.2 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr8_-_62559366 | 0.15 |
ENST00000522919.1 |
ASPH |
aspartate beta-hydroxylase |
| chr15_-_78913628 | 0.15 |
ENST00000348639.3 |
CHRNA3 |
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr12_+_75874984 | 0.15 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_-_43833515 | 0.15 |
ENST00000549670.1 ENST00000395541.2 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr10_-_79398127 | 0.15 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_74606223 | 0.15 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr4_-_76928641 | 0.15 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr17_-_8113542 | 0.15 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr19_+_50016610 | 0.15 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr8_-_117043 | 0.15 |
ENST00000320901.3 |
OR4F21 |
olfactory receptor, family 4, subfamily F, member 21 |
| chr14_+_52164820 | 0.15 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr5_-_96518907 | 0.14 |
ENST00000508447.1 ENST00000283109.3 |
RIOK2 |
RIO kinase 2 |
| chr12_-_92536433 | 0.14 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr14_-_81425828 | 0.14 |
ENST00000555529.1 ENST00000556042.1 ENST00000556981.1 |
CEP128 |
centrosomal protein 128kDa |
| chr1_+_87012753 | 0.14 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr11_-_102576537 | 0.14 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr5_+_112073544 | 0.14 |
ENST00000257430.4 ENST00000508376.2 |
APC |
adenomatous polyposis coli |
| chr15_+_65843130 | 0.14 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr7_-_150020578 | 0.14 |
ENST00000478393.1 |
ACTR3C |
ARP3 actin-related protein 3 homolog C (yeast) |
| chr1_+_197170592 | 0.14 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr2_+_152214098 | 0.14 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr12_+_75874460 | 0.14 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr13_-_24007815 | 0.14 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr18_-_33702078 | 0.14 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr12_-_89920030 | 0.14 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr1_+_197237352 | 0.13 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr12_-_10151773 | 0.13 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr1_+_115572415 | 0.13 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr2_-_111334678 | 0.13 |
ENST00000329516.3 ENST00000330331.5 ENST00000446930.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr20_+_34802295 | 0.13 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr5_+_129083772 | 0.13 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr4_-_70626430 | 0.13 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr12_-_11287243 | 0.13 |
ENST00000539585.1 |
TAS2R30 |
taste receptor, type 2, member 30 |
| chrX_+_77166172 | 0.13 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr18_-_14132422 | 0.13 |
ENST00000589498.1 ENST00000590202.1 |
ZNF519 |
zinc finger protein 519 |
| chr1_-_232651312 | 0.13 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr1_-_183622442 | 0.13 |
ENST00000308641.4 |
APOBEC4 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr5_-_89770582 | 0.13 |
ENST00000316610.6 |
MBLAC2 |
metallo-beta-lactamase domain containing 2 |
| chr2_-_89340242 | 0.12 |
ENST00000480492.1 |
IGKV1-12 |
immunoglobulin kappa variable 1-12 |
| chr21_+_39644305 | 0.12 |
ENST00000398930.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_24540046 | 0.12 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr11_+_12766583 | 0.12 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr11_-_111649015 | 0.12 |
ENST00000529841.1 |
RP11-108O10.2 |
RP11-108O10.2 |
| chr8_+_27632083 | 0.12 |
ENST00000519637.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_-_60539422 | 0.12 |
ENST00000371201.3 |
C1orf87 |
chromosome 1 open reading frame 87 |
| chr22_+_39966758 | 0.12 |
ENST00000407673.1 ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr15_+_65822756 | 0.12 |
ENST00000562901.1 ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr2_+_197577841 | 0.12 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr11_-_27722021 | 0.12 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chr20_-_56285595 | 0.12 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr9_+_120466650 | 0.12 |
ENST00000355622.6 |
TLR4 |
toll-like receptor 4 |
| chr19_-_22034809 | 0.12 |
ENST00000594012.1 ENST00000595461.1 ENST00000596899.1 |
ZNF43 |
zinc finger protein 43 |
| chr5_+_140480083 | 0.12 |
ENST00000231130.2 |
PCDHB3 |
protocadherin beta 3 |
| chr2_+_228736335 | 0.12 |
ENST00000440997.1 ENST00000545118.1 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr1_-_67266939 | 0.12 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr12_+_75874580 | 0.12 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr11_-_59633951 | 0.12 |
ENST00000257264.3 |
TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr2_+_11864458 | 0.12 |
ENST00000396098.1 ENST00000396099.1 ENST00000425416.2 |
LPIN1 |
lipin 1 |
| chr15_+_90895471 | 0.12 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chr17_-_67264947 | 0.12 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr1_-_211307404 | 0.12 |
ENST00000367007.4 |
KCNH1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr15_+_41913690 | 0.12 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr19_+_52873166 | 0.12 |
ENST00000424032.2 ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880 |
zinc finger protein 880 |
| chr12_+_60083118 | 0.11 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr15_+_69365265 | 0.11 |
ENST00000415504.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr6_+_52442083 | 0.11 |
ENST00000606714.1 |
TRAM2-AS1 |
TRAM2 antisense RNA 1 (head to head) |
| chr11_+_30253410 | 0.11 |
ENST00000533718.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr2_+_169757750 | 0.11 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr12_-_7596735 | 0.11 |
ENST00000416109.2 ENST00000396630.1 ENST00000313599.3 |
CD163L1 |
CD163 molecule-like 1 |
| chr15_+_89631381 | 0.11 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr19_+_45449301 | 0.11 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr7_-_47579188 | 0.11 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr3_-_149093499 | 0.11 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr6_+_28092338 | 0.11 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr11_+_35201826 | 0.11 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr11_-_5323226 | 0.11 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr6_+_140175987 | 0.11 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr8_+_66955648 | 0.11 |
ENST00000522619.1 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr9_-_73477826 | 0.11 |
ENST00000396285.1 ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_-_58459039 | 0.11 |
ENST00000282308.3 ENST00000598928.1 |
ZNF256 |
zinc finger protein 256 |
| chr10_+_133747955 | 0.11 |
ENST00000455566.1 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr14_-_106539557 | 0.11 |
ENST00000390599.2 |
IGHV1-8 |
immunoglobulin heavy variable 1-8 |
| chr13_+_24144796 | 0.11 |
ENST00000403372.2 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr12_+_59989918 | 0.11 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_+_17281900 | 0.11 |
ENST00000530527.1 |
NUCB2 |
nucleobindin 2 |
| chr2_+_109237717 | 0.11 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_-_32908792 | 0.11 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr16_+_22516172 | 0.11 |
ENST00000543407.1 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr4_-_66536057 | 0.11 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr11_-_69633792 | 0.11 |
ENST00000334134.2 |
FGF3 |
fibroblast growth factor 3 |
| chr13_+_107029084 | 0.11 |
ENST00000444865.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr1_+_155829286 | 0.11 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr1_-_186344802 | 0.11 |
ENST00000451586.1 |
TPR |
translocated promoter region, nuclear basket protein |
| chr16_+_15489603 | 0.11 |
ENST00000568766.1 ENST00000287594.7 |
RP11-1021N1.1 MPV17L |
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr6_+_29274403 | 0.10 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr17_+_34431212 | 0.10 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr1_-_106161540 | 0.10 |
ENST00000420901.1 ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1 |
RP11-251P6.1 |
| chr10_+_57358750 | 0.10 |
ENST00000512524.2 |
MTRNR2L5 |
MT-RNR2-like 5 |
| chr14_+_60558627 | 0.10 |
ENST00000317623.4 ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4 |
pecanex-like 4 (Drosophila) |
| chr12_+_32260085 | 0.10 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr1_-_232697304 | 0.10 |
ENST00000366630.1 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr2_+_238395803 | 0.10 |
ENST00000264605.3 |
MLPH |
melanophilin |
| chrX_+_44703249 | 0.10 |
ENST00000339042.4 |
DUSP21 |
dual specificity phosphatase 21 |
| chr4_+_110749143 | 0.10 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr3_+_141103634 | 0.10 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr11_+_125462690 | 0.10 |
ENST00000392708.4 ENST00000529196.1 ENST00000531491.1 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr12_+_15475462 | 0.10 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr13_+_50570019 | 0.10 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr16_+_20499024 | 0.10 |
ENST00000593357.1 |
AC137056.1 |
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr10_-_14996321 | 0.10 |
ENST00000378289.4 |
DCLRE1C |
DNA cross-link repair 1C |
| chr2_-_145275228 | 0.10 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr7_+_129932974 | 0.10 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr3_-_168865522 | 0.10 |
ENST00000464456.1 |
MECOM |
MDS1 and EVI1 complex locus |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.9 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.2 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.0 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.0 | GO:1902956 | regulation of oxidative phosphorylation(GO:0002082) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.0 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.6 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.0 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.0 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0005217 | intracellular ligand-gated ion channel activity(GO:0005217) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.0 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |