Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for DPRX
Z-value: 1.34
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | DPRX |
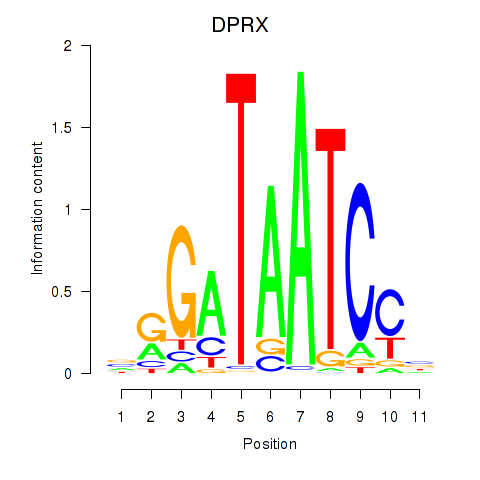
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_105845674 | 1.66 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr1_+_35220613 | 1.56 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr10_+_118187379 | 1.52 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr18_-_28681950 | 1.44 |
ENST00000251081.6 |
DSC2 |
desmocollin 2 |
| chr5_-_150948414 | 1.33 |
ENST00000261800.5 |
FAT2 |
FAT atypical cadherin 2 |
| chr9_-_23821273 | 1.21 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr18_-_28682374 | 1.15 |
ENST00000280904.6 |
DSC2 |
desmocollin 2 |
| chr3_+_136676851 | 1.08 |
ENST00000309741.5 |
IL20RB |
interleukin 20 receptor beta |
| chr17_-_4643114 | 1.04 |
ENST00000293778.6 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
| chr3_+_136676707 | 1.01 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr2_-_216003127 | 0.94 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr2_-_235405168 | 0.93 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr1_+_81771806 | 0.87 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr14_-_22005343 | 0.82 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr18_-_47376197 | 0.81 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr12_+_113344755 | 0.77 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr14_-_22005062 | 0.76 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chrX_-_33229636 | 0.75 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr14_-_22005018 | 0.73 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr5_+_35856951 | 0.72 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr6_-_13487784 | 0.71 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr4_-_170924888 | 0.69 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr14_+_61995722 | 0.61 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr5_+_140345820 | 0.60 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr6_-_32784687 | 0.60 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr11_-_104916034 | 0.58 |
ENST00000528513.1 ENST00000375706.2 ENST00000375704.3 |
CARD16 |
caspase recruitment domain family, member 16 |
| chr1_-_153521597 | 0.58 |
ENST00000368712.1 |
S100A3 |
S100 calcium binding protein A3 |
| chr1_-_153521714 | 0.56 |
ENST00000368713.3 |
S100A3 |
S100 calcium binding protein A3 |
| chr18_-_52989217 | 0.56 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr3_-_112564797 | 0.55 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr8_-_80993010 | 0.53 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr4_+_69313145 | 0.52 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr18_-_52989525 | 0.52 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr17_-_9694614 | 0.51 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr4_+_89299885 | 0.49 |
ENST00000380265.5 ENST00000273960.3 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_+_113344582 | 0.47 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_6949964 | 0.47 |
ENST00000541978.1 ENST00000435982.2 |
GNB3 |
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr11_+_61248583 | 0.46 |
ENST00000432063.2 ENST00000338608.2 |
PPP1R32 |
protein phosphatase 1, regulatory subunit 32 |
| chrX_-_15683147 | 0.45 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr2_+_103378472 | 0.45 |
ENST00000412401.2 |
TMEM182 |
transmembrane protein 182 |
| chr8_-_95274536 | 0.44 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr7_+_30323923 | 0.44 |
ENST00000323037.4 |
ZNRF2 |
zinc and ring finger 2 |
| chr11_-_104905840 | 0.44 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr14_+_70918874 | 0.43 |
ENST00000603540.1 |
ADAM21 |
ADAM metallopeptidase domain 21 |
| chr11_-_104972158 | 0.42 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_-_170599561 | 0.42 |
ENST00000366756.3 |
DLL1 |
delta-like 1 (Drosophila) |
| chr11_-_104817919 | 0.42 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr12_+_113344811 | 0.42 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr6_+_130339710 | 0.40 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr1_-_68516393 | 0.40 |
ENST00000395201.1 |
DIRAS3 |
DIRAS family, GTP-binding RAS-like 3 |
| chr11_-_321050 | 0.39 |
ENST00000399808.4 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr16_-_70719925 | 0.39 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr12_+_133066137 | 0.39 |
ENST00000434748.2 |
FBRSL1 |
fibrosin-like 1 |
| chr6_-_13487825 | 0.39 |
ENST00000603223.1 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr14_+_22670455 | 0.38 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chrX_+_99899180 | 0.38 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr4_-_87374283 | 0.37 |
ENST00000361569.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr20_+_361890 | 0.37 |
ENST00000449710.1 ENST00000422053.2 |
TRIB3 |
tribbles pseudokinase 3 |
| chr22_-_19137796 | 0.35 |
ENST00000086933.2 |
GSC2 |
goosecoid homeobox 2 |
| chr19_+_41222998 | 0.35 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr22_+_32754139 | 0.34 |
ENST00000382088.3 |
RFPL3 |
ret finger protein-like 3 |
| chr14_+_91581011 | 0.34 |
ENST00000523894.1 ENST00000522322.1 ENST00000523771.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr6_-_40555176 | 0.32 |
ENST00000338305.6 |
LRFN2 |
leucine rich repeat and fibronectin type III domain containing 2 |
| chr20_+_1875942 | 0.32 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr17_+_55183261 | 0.31 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr6_+_37225540 | 0.31 |
ENST00000373491.3 |
TBC1D22B |
TBC1 domain family, member 22B |
| chr6_-_35480640 | 0.30 |
ENST00000428978.1 ENST00000322263.4 |
TULP1 |
tubby like protein 1 |
| chr1_-_184943610 | 0.29 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr11_-_124190184 | 0.29 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr18_-_64271363 | 0.28 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr11_-_321340 | 0.28 |
ENST00000526811.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr1_+_111415757 | 0.27 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr1_+_156117149 | 0.27 |
ENST00000435124.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr4_+_159131346 | 0.27 |
ENST00000508243.1 ENST00000296529.6 |
TMEM144 |
transmembrane protein 144 |
| chr1_-_22263790 | 0.26 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr4_-_123542224 | 0.25 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr3_+_111630451 | 0.24 |
ENST00000495180.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr18_-_64271316 | 0.24 |
ENST00000540086.1 ENST00000580157.1 |
CDH19 |
cadherin 19, type 2 |
| chr18_+_56530136 | 0.24 |
ENST00000591083.1 |
ZNF532 |
zinc finger protein 532 |
| chr19_-_4831701 | 0.24 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr16_-_4665023 | 0.23 |
ENST00000591897.1 |
UBALD1 |
UBA-like domain containing 1 |
| chr6_+_7107999 | 0.23 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr12_+_130554803 | 0.23 |
ENST00000535487.1 |
RP11-474D1.2 |
RP11-474D1.2 |
| chr4_+_147096837 | 0.23 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_+_4643300 | 0.23 |
ENST00000433935.1 |
ZMYND15 |
zinc finger, MYND-type containing 15 |
| chr22_+_23161491 | 0.23 |
ENST00000390316.2 |
IGLV3-9 |
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chrX_+_15767971 | 0.22 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr1_-_161014731 | 0.22 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chrX_+_15525426 | 0.21 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chrX_-_3631635 | 0.21 |
ENST00000262848.5 |
PRKX |
protein kinase, X-linked |
| chr8_-_110656995 | 0.21 |
ENST00000276646.9 ENST00000533065.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr19_+_8455077 | 0.20 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr16_-_4664860 | 0.20 |
ENST00000587615.1 ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1 |
UBA-like domain containing 1 |
| chr7_+_123241908 | 0.20 |
ENST00000434204.1 ENST00000437535.1 ENST00000451215.1 |
ASB15 |
ankyrin repeat and SOCS box containing 15 |
| chr8_+_145597713 | 0.20 |
ENST00000308860.6 ENST00000532190.1 |
ADCK5 |
aarF domain containing kinase 5 |
| chr17_+_4643337 | 0.19 |
ENST00000592813.1 |
ZMYND15 |
zinc finger, MYND-type containing 15 |
| chr2_+_33701286 | 0.19 |
ENST00000403687.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_+_7531281 | 0.19 |
ENST00000575729.1 ENST00000340624.5 |
SHBG |
sex hormone-binding globulin |
| chr2_+_113342011 | 0.18 |
ENST00000324913.5 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr16_-_20556492 | 0.18 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr11_+_64949158 | 0.18 |
ENST00000527739.1 ENST00000526966.1 ENST00000533129.1 ENST00000524773.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr2_+_26915584 | 0.18 |
ENST00000302909.3 |
KCNK3 |
potassium channel, subfamily K, member 3 |
| chr16_+_21244986 | 0.18 |
ENST00000311620.5 |
ANKS4B |
ankyrin repeat and sterile alpha motif domain containing 4B |
| chrX_-_77150911 | 0.18 |
ENST00000373336.3 |
MAGT1 |
magnesium transporter 1 |
| chr2_+_113342163 | 0.18 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr2_-_26700900 | 0.18 |
ENST00000338581.6 ENST00000339598.3 ENST00000402415.3 |
OTOF |
otoferlin |
| chr17_-_1928621 | 0.18 |
ENST00000331238.6 |
RTN4RL1 |
reticulon 4 receptor-like 1 |
| chr8_-_124037890 | 0.17 |
ENST00000519018.1 ENST00000523036.1 |
DERL1 |
derlin 1 |
| chr17_-_19771242 | 0.17 |
ENST00000361658.2 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr1_-_55089191 | 0.17 |
ENST00000302250.2 ENST00000371304.2 |
FAM151A |
family with sequence similarity 151, member A |
| chr10_+_74451883 | 0.17 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chr3_+_97483366 | 0.16 |
ENST00000463745.1 ENST00000462412.1 |
ARL6 |
ADP-ribosylation factor-like 6 |
| chr11_+_2323236 | 0.16 |
ENST00000182290.4 |
TSPAN32 |
tetraspanin 32 |
| chr19_-_7040190 | 0.16 |
ENST00000381394.4 |
MBD3L4 |
methyl-CpG binding domain protein 3-like 4 |
| chr5_-_37835929 | 0.16 |
ENST00000381826.4 ENST00000427982.1 |
GDNF |
glial cell derived neurotrophic factor |
| chr6_-_35480705 | 0.16 |
ENST00000229771.6 |
TULP1 |
tubby like protein 1 |
| chr19_+_7030589 | 0.16 |
ENST00000329753.5 |
MBD3L5 |
methyl-CpG binding domain protein 3-like 5 |
| chr3_+_101659682 | 0.16 |
ENST00000465215.1 |
RP11-221J22.1 |
RP11-221J22.1 |
| chr19_-_50370799 | 0.16 |
ENST00000600910.1 ENST00000322344.3 ENST00000600573.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr17_-_4448361 | 0.16 |
ENST00000572759.1 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chr5_+_180336564 | 0.15 |
ENST00000505126.1 ENST00000533815.2 |
BTNL8 |
butyrophilin-like 8 |
| chr9_+_116207007 | 0.14 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr2_-_49381572 | 0.14 |
ENST00000454032.1 ENST00000304421.4 |
FSHR |
follicle stimulating hormone receptor |
| chr16_-_50913188 | 0.14 |
ENST00000569986.1 |
CTD-2034I21.1 |
CTD-2034I21.1 |
| chr3_+_15468862 | 0.14 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr2_-_49381646 | 0.14 |
ENST00000346173.3 ENST00000406846.2 |
FSHR |
follicle stimulating hormone receptor |
| chr2_+_232063436 | 0.14 |
ENST00000440107.1 |
ARMC9 |
armadillo repeat containing 9 |
| chr1_+_247582097 | 0.14 |
ENST00000391827.2 |
NLRP3 |
NLR family, pyrin domain containing 3 |
| chr11_-_85430204 | 0.14 |
ENST00000389958.3 ENST00000527794.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr17_-_26127525 | 0.14 |
ENST00000313735.6 |
NOS2 |
nitric oxide synthase 2, inducible |
| chr9_+_4839762 | 0.14 |
ENST00000448872.2 ENST00000441844.1 |
RCL1 |
RNA terminal phosphate cyclase-like 1 |
| chr11_-_85430163 | 0.14 |
ENST00000529581.1 ENST00000533577.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr12_-_104359475 | 0.14 |
ENST00000553183.1 |
C12orf73 |
chromosome 12 open reading frame 73 |
| chr19_-_7058651 | 0.13 |
ENST00000333843.4 |
MBD3L3 |
methyl-CpG binding domain protein 3-like 3 |
| chr7_-_135412925 | 0.13 |
ENST00000354042.4 |
SLC13A4 |
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr7_-_99869799 | 0.13 |
ENST00000436886.2 |
GATS |
GATS, stromal antigen 3 opposite strand |
| chr7_-_155601766 | 0.13 |
ENST00000430104.1 |
SHH |
sonic hedgehog |
| chr2_+_114737146 | 0.13 |
ENST00000412690.1 |
AC010982.1 |
AC010982.1 |
| chr16_-_46723066 | 0.13 |
ENST00000299138.7 |
VPS35 |
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr1_-_13452656 | 0.13 |
ENST00000376132.3 |
PRAMEF13 |
PRAME family member 13 |
| chr1_-_94586651 | 0.13 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr19_+_7049332 | 0.13 |
ENST00000381393.3 |
MBD3L2 |
methyl-CpG binding domain protein 3-like 2 |
| chr6_+_26383404 | 0.13 |
ENST00000416795.2 ENST00000494184.1 |
BTN2A2 |
butyrophilin, subfamily 2, member A2 |
| chr4_-_153700864 | 0.12 |
ENST00000304337.2 |
TIGD4 |
tigger transposable element derived 4 |
| chr11_-_77122928 | 0.12 |
ENST00000528203.1 ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr1_-_36615065 | 0.12 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr7_+_135242652 | 0.12 |
ENST00000285968.6 ENST00000440390.2 |
NUP205 |
nucleoporin 205kDa |
| chr6_+_4706368 | 0.12 |
ENST00000328908.5 |
CDYL |
chromodomain protein, Y-like |
| chr8_-_117886955 | 0.12 |
ENST00000297338.2 |
RAD21 |
RAD21 homolog (S. pombe) |
| chr5_-_77072085 | 0.12 |
ENST00000518338.2 ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA |
tubulin folding cofactor A |
| chr2_+_234216454 | 0.12 |
ENST00000447536.1 ENST00000409110.1 |
SAG |
S-antigen; retina and pineal gland (arrestin) |
| chr18_-_3845293 | 0.12 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr8_+_8559406 | 0.12 |
ENST00000519106.1 |
CLDN23 |
claudin 23 |
| chr4_-_47465666 | 0.11 |
ENST00000381571.4 |
COMMD8 |
COMM domain containing 8 |
| chr2_+_145425573 | 0.11 |
ENST00000600064.1 ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr19_+_36157715 | 0.11 |
ENST00000379013.2 ENST00000222275.2 |
UPK1A |
uroplakin 1A |
| chr11_+_107879459 | 0.11 |
ENST00000393094.2 |
CUL5 |
cullin 5 |
| chr8_+_145215928 | 0.10 |
ENST00000528919.1 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr1_-_27481401 | 0.10 |
ENST00000263980.3 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr6_-_132967142 | 0.10 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chr1_+_43148059 | 0.10 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr1_+_236557569 | 0.10 |
ENST00000334232.4 |
EDARADD |
EDAR-associated death domain |
| chr2_-_119605253 | 0.10 |
ENST00000295206.6 |
EN1 |
engrailed homeobox 1 |
| chr3_+_23847432 | 0.10 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr5_-_142065223 | 0.10 |
ENST00000378046.1 |
FGF1 |
fibroblast growth factor 1 (acidic) |
| chr6_+_26383318 | 0.10 |
ENST00000469230.1 ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2 |
butyrophilin, subfamily 2, member A2 |
| chr1_-_170043709 | 0.10 |
ENST00000367767.1 ENST00000361580.2 ENST00000538366.1 |
KIFAP3 |
kinesin-associated protein 3 |
| chr11_-_62752162 | 0.10 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr19_+_36139125 | 0.10 |
ENST00000246554.3 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr2_+_33661382 | 0.10 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_-_19771216 | 0.10 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr6_-_128239749 | 0.10 |
ENST00000537166.1 |
THEMIS |
thymocyte selection associated |
| chr2_-_68290106 | 0.10 |
ENST00000407324.1 ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D |
C1D nuclear receptor corepressor |
| chr4_-_103997862 | 0.09 |
ENST00000394785.3 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr10_-_89577910 | 0.09 |
ENST00000308448.7 ENST00000541004.1 |
ATAD1 |
ATPase family, AAA domain containing 1 |
| chr6_-_128239685 | 0.09 |
ENST00000368250.1 |
THEMIS |
thymocyte selection associated |
| chr17_-_3182268 | 0.09 |
ENST00000408891.2 |
OR3A2 |
olfactory receptor, family 3, subfamily A, member 2 |
| chr19_+_48325097 | 0.09 |
ENST00000221996.7 ENST00000539067.1 |
CRX |
cone-rod homeobox |
| chr2_-_98280383 | 0.09 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr4_+_113970772 | 0.09 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr5_-_77072145 | 0.09 |
ENST00000380377.4 |
TBCA |
tubulin folding cofactor A |
| chr1_+_13516066 | 0.09 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr2_+_145425534 | 0.09 |
ENST00000432608.1 ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr17_-_48785216 | 0.08 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr4_+_159131630 | 0.08 |
ENST00000504569.1 ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144 |
transmembrane protein 144 |
| chr4_-_100356551 | 0.08 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_-_45696253 | 0.08 |
ENST00000303230.4 |
HCN1 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr11_-_85430088 | 0.08 |
ENST00000533057.1 ENST00000533892.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr19_-_14628645 | 0.08 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr12_-_71182695 | 0.08 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr17_+_41150290 | 0.08 |
ENST00000589037.1 ENST00000253788.5 |
RPL27 |
ribosomal protein L27 |
| chr4_-_73935409 | 0.08 |
ENST00000507544.2 ENST00000295890.4 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
| chr2_+_74685413 | 0.08 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr1_-_173174681 | 0.08 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr6_+_7108210 | 0.08 |
ENST00000467782.1 ENST00000334984.6 ENST00000349384.6 |
RREB1 |
ras responsive element binding protein 1 |
| chr1_+_20396649 | 0.08 |
ENST00000375108.3 |
PLA2G5 |
phospholipase A2, group V |
| chr8_+_65285851 | 0.08 |
ENST00000520799.1 ENST00000521441.1 |
LINC00966 |
long intergenic non-protein coding RNA 966 |
| chr3_+_132757215 | 0.08 |
ENST00000321871.6 ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108 |
transmembrane protein 108 |
| chr3_+_40351169 | 0.07 |
ENST00000232905.3 |
EIF1B |
eukaryotic translation initiation factor 1B |
| chrX_+_118425471 | 0.07 |
ENST00000428222.1 |
RP5-1139I1.1 |
RP5-1139I1.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.4 | 1.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 2.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 0.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.4 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.0 | 0.1 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.9 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 2.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.0 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.4 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 2.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 2.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 0.9 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.2 | 1.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |