Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for E2F8
Z-value: 0.48
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19262486_19262512 | -0.47 | 2.4e-01 | Click! |
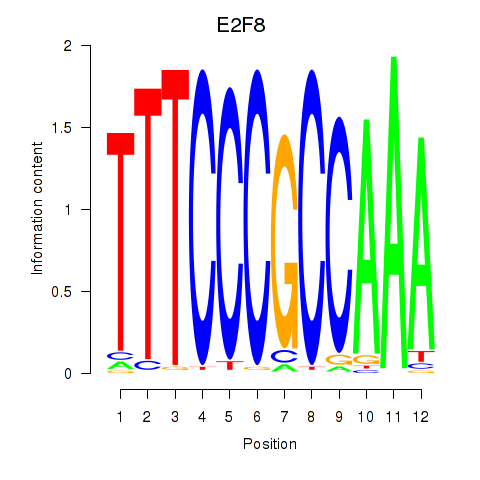
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_17434689 | 0.68 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chrX_+_51927919 | 0.57 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr2_+_24346324 | 0.49 |
ENST00000407625.1 ENST00000420135.2 |
FAM228B |
family with sequence similarity 228, member B |
| chr20_-_34025999 | 0.41 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr10_+_70847852 | 0.39 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr9_+_99690592 | 0.35 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr5_+_82767583 | 0.35 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr5_+_82767487 | 0.32 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr12_+_1738363 | 0.31 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr2_-_24346218 | 0.27 |
ENST00000436622.1 ENST00000313213.4 |
PFN4 |
profilin family, member 4 |
| chrX_+_51928002 | 0.27 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr10_+_104178946 | 0.25 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr1_+_197871854 | 0.25 |
ENST00000436652.1 |
C1orf53 |
chromosome 1 open reading frame 53 |
| chrX_-_51812268 | 0.25 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr2_+_238395803 | 0.23 |
ENST00000264605.3 |
MLPH |
melanophilin |
| chr2_+_238395879 | 0.23 |
ENST00000445024.2 ENST00000338530.4 ENST00000409373.1 |
MLPH |
melanophilin |
| chr1_+_197871740 | 0.23 |
ENST00000367393.3 |
C1orf53 |
chromosome 1 open reading frame 53 |
| chr13_-_43566301 | 0.22 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr14_-_74551096 | 0.21 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr19_-_51845378 | 0.21 |
ENST00000335624.4 |
VSIG10L |
V-set and immunoglobulin domain containing 10 like |
| chr2_+_148778570 | 0.21 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr12_-_15942309 | 0.21 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr3_-_66551351 | 0.20 |
ENST00000273261.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr12_-_15942503 | 0.20 |
ENST00000281172.5 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr17_-_15165854 | 0.20 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr3_-_66551397 | 0.19 |
ENST00000383703.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr1_+_156024552 | 0.18 |
ENST00000368304.5 ENST00000368302.3 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr11_+_71938925 | 0.18 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr9_+_124088860 | 0.18 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr1_+_150245177 | 0.17 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr12_+_107168418 | 0.17 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr14_-_74551172 | 0.16 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr15_-_89456630 | 0.15 |
ENST00000268150.8 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr1_+_156024525 | 0.15 |
ENST00000368305.4 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr15_-_89456593 | 0.15 |
ENST00000558029.1 ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr1_+_150245099 | 0.15 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr22_+_42086547 | 0.14 |
ENST00000402966.1 |
C22orf46 |
chromosome 22 open reading frame 46 |
| chr8_-_7243080 | 0.14 |
ENST00000400156.4 |
ZNF705G |
zinc finger protein 705G |
| chr6_-_57086200 | 0.14 |
ENST00000468148.1 |
RAB23 |
RAB23, member RAS oncogene family |
| chrX_+_150866779 | 0.14 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr11_-_66445219 | 0.14 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr17_+_39975455 | 0.14 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr5_+_172483347 | 0.13 |
ENST00000522692.1 ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF |
CREB3 regulatory factor |
| chr17_+_47075023 | 0.13 |
ENST00000431824.2 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_+_41103063 | 0.13 |
ENST00000308370.7 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr19_+_49977466 | 0.12 |
ENST00000596435.1 ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr12_+_107168342 | 0.12 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr19_+_49977818 | 0.12 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr15_+_44719970 | 0.12 |
ENST00000558966.1 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr15_+_44119159 | 0.12 |
ENST00000263795.6 ENST00000381246.2 ENST00000452115.1 |
WDR76 |
WD repeat domain 76 |
| chr12_+_18414446 | 0.12 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr17_+_39975544 | 0.12 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr17_+_75401152 | 0.12 |
ENST00000585930.1 |
SEPT9 |
septin 9 |
| chr11_+_118992269 | 0.12 |
ENST00000350777.2 ENST00000529988.1 ENST00000527410.1 |
HINFP |
histone H4 transcription factor |
| chr12_-_6772303 | 0.12 |
ENST00000396807.4 ENST00000446105.2 ENST00000341550.4 |
ING4 |
inhibitor of growth family, member 4 |
| chr15_-_57025759 | 0.12 |
ENST00000267807.7 |
ZNF280D |
zinc finger protein 280D |
| chrX_-_153707545 | 0.12 |
ENST00000357360.4 |
LAGE3 |
L antigen family, member 3 |
| chr12_-_102455846 | 0.11 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr12_-_54071181 | 0.11 |
ENST00000338662.5 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chrX_-_153707246 | 0.11 |
ENST00000407062.1 |
LAGE3 |
L antigen family, member 3 |
| chr1_-_17307173 | 0.11 |
ENST00000438542.1 ENST00000375535.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr3_-_49466686 | 0.11 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr6_-_85473156 | 0.11 |
ENST00000606784.1 ENST00000606325.1 |
TBX18 |
T-box 18 |
| chr1_-_159893507 | 0.10 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chr12_-_102455902 | 0.10 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr15_-_81616446 | 0.10 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr1_-_150693318 | 0.10 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr12_+_56661461 | 0.10 |
ENST00000546544.1 ENST00000553234.1 |
COQ10A |
coenzyme Q10 homolog A (S. cerevisiae) |
| chr1_-_36916066 | 0.10 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr16_-_67185117 | 0.10 |
ENST00000449549.3 |
B3GNT9 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr2_-_152146385 | 0.10 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr14_-_24898731 | 0.10 |
ENST00000267406.6 |
CBLN3 |
cerebellin 3 precursor |
| chr10_-_74385876 | 0.10 |
ENST00000398761.4 |
MICU1 |
mitochondrial calcium uptake 1 |
| chr12_-_48499591 | 0.10 |
ENST00000551330.1 ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1 |
SUMO1/sentrin specific peptidase 1 |
| chr4_-_21699380 | 0.10 |
ENST00000382148.3 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr16_-_30134266 | 0.09 |
ENST00000484663.1 ENST00000478356.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr20_-_39946237 | 0.09 |
ENST00000441102.2 ENST00000559234.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr15_+_64428529 | 0.09 |
ENST00000560861.1 |
SNX1 |
sorting nexin 1 |
| chr10_-_74385811 | 0.09 |
ENST00000603011.1 ENST00000401998.3 ENST00000361114.5 ENST00000604238.1 |
MICU1 |
mitochondrial calcium uptake 1 |
| chr6_-_2962331 | 0.09 |
ENST00000380524.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr17_+_66243715 | 0.09 |
ENST00000359904.3 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr3_+_69985792 | 0.09 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr17_-_9479128 | 0.09 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr1_+_97187318 | 0.09 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr8_-_92053212 | 0.09 |
ENST00000285419.3 |
TMEM55A |
transmembrane protein 55A |
| chr1_-_26232522 | 0.09 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr6_-_111804905 | 0.09 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr18_+_72922710 | 0.08 |
ENST00000322038.5 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
| chr10_+_35416090 | 0.08 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr10_+_124670121 | 0.08 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr1_+_244998918 | 0.08 |
ENST00000366528.3 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr18_-_12656715 | 0.08 |
ENST00000462226.1 ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1 |
spire-type actin nucleation factor 1 |
| chr9_-_123639600 | 0.08 |
ENST00000373896.3 |
PHF19 |
PHD finger protein 19 |
| chr14_-_61190754 | 0.08 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr14_+_55595762 | 0.08 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr17_+_57642886 | 0.08 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr6_+_127588020 | 0.08 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr3_+_14058794 | 0.08 |
ENST00000424053.1 ENST00000528067.1 ENST00000429201.1 |
TPRXL |
tetra-peptide repeat homeobox-like |
| chr7_-_56118981 | 0.07 |
ENST00000419984.2 ENST00000413218.1 ENST00000424596.1 |
PSPH |
phosphoserine phosphatase |
| chr10_-_104178857 | 0.07 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr4_-_129209944 | 0.07 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr11_+_47236489 | 0.07 |
ENST00000256996.4 ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2 |
damage-specific DNA binding protein 2, 48kDa |
| chr11_+_33061543 | 0.07 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr1_-_36916011 | 0.07 |
ENST00000356637.5 ENST00000354267.3 ENST00000235532.5 |
OSCP1 |
organic solute carrier partner 1 |
| chr1_+_170501270 | 0.07 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr14_+_77582905 | 0.07 |
ENST00000557408.1 |
TMEM63C |
transmembrane protein 63C |
| chr11_-_108369101 | 0.07 |
ENST00000323468.5 |
KDELC2 |
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr9_-_123639304 | 0.07 |
ENST00000436309.1 |
PHF19 |
PHD finger protein 19 |
| chr20_+_31595406 | 0.07 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr2_+_130939235 | 0.07 |
ENST00000425361.1 ENST00000457492.1 |
MZT2B |
mitotic spindle organizing protein 2B |
| chr14_-_101036119 | 0.07 |
ENST00000355173.2 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr2_+_231191875 | 0.07 |
ENST00000444636.1 ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L |
SP140 nuclear body protein-like |
| chr10_-_17659234 | 0.07 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chrX_+_52240504 | 0.07 |
ENST00000399805.2 |
XAGE1B |
X antigen family, member 1B |
| chr7_-_56119156 | 0.06 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr7_-_21985656 | 0.06 |
ENST00000406877.3 |
CDCA7L |
cell division cycle associated 7-like |
| chr1_-_205719295 | 0.06 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr1_+_39876151 | 0.06 |
ENST00000530275.1 |
KIAA0754 |
KIAA0754 |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr22_-_36877629 | 0.06 |
ENST00000416967.1 |
TXN2 |
thioredoxin 2 |
| chr19_+_12203069 | 0.06 |
ENST00000430298.2 ENST00000339302.4 |
ZNF788 ZNF788 |
zinc finger family member 788 Zinc finger protein 788 |
| chr2_+_79252804 | 0.06 |
ENST00000393897.2 |
REG3G |
regenerating islet-derived 3 gamma |
| chr14_+_79745682 | 0.06 |
ENST00000557594.1 |
NRXN3 |
neurexin 3 |
| chr7_-_21985489 | 0.06 |
ENST00000356195.5 ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L |
cell division cycle associated 7-like |
| chr22_+_35796108 | 0.06 |
ENST00000382011.5 ENST00000416905.1 |
MCM5 |
minichromosome maintenance complex component 5 |
| chr2_+_79252822 | 0.06 |
ENST00000272324.5 |
REG3G |
regenerating islet-derived 3 gamma |
| chr12_-_110939870 | 0.06 |
ENST00000447578.2 ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29 |
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr14_+_24837226 | 0.06 |
ENST00000554050.1 ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr17_-_61819121 | 0.06 |
ENST00000245865.5 ENST00000375840.4 ENST00000582137.1 ENST00000579549.1 ENST00000582030.1 ENST00000584110.1 ENST00000580288.1 ENST00000336174.6 ENST00000579340.1 ENST00000580338.1 |
STRADA |
STE20-related kinase adaptor alpha |
| chr5_+_142125161 | 0.06 |
ENST00000432677.1 |
AC005592.1 |
AC005592.1 |
| chr18_+_5238055 | 0.06 |
ENST00000582363.1 ENST00000582008.1 ENST00000580082.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr14_-_69619291 | 0.05 |
ENST00000554215.1 ENST00000556847.1 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr19_+_16296191 | 0.05 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr22_+_38071615 | 0.05 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr4_+_108910870 | 0.05 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr19_+_36132631 | 0.05 |
ENST00000379026.2 ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2 |
ets variant 2 |
| chr16_+_70332956 | 0.05 |
ENST00000288071.6 ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B RP11-529K1.3 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr14_-_69619823 | 0.05 |
ENST00000341516.5 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr3_-_146187088 | 0.05 |
ENST00000497985.1 |
PLSCR2 |
phospholipid scramblase 2 |
| chr2_+_112939365 | 0.05 |
ENST00000272559.4 |
FBLN7 |
fibulin 7 |
| chr19_-_12267524 | 0.05 |
ENST00000455799.1 ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625 |
zinc finger protein 625 |
| chr17_+_61904766 | 0.05 |
ENST00000581842.1 ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr15_-_77712477 | 0.05 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr3_+_12598563 | 0.05 |
ENST00000411987.1 ENST00000448482.1 |
MKRN2 |
makorin ring finger protein 2 |
| chr6_-_84937314 | 0.05 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr4_-_140223670 | 0.05 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_63815628 | 0.05 |
ENST00000409562.3 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr2_-_198650037 | 0.05 |
ENST00000392296.4 |
BOLL |
boule-like RNA-binding protein |
| chr11_+_118868830 | 0.05 |
ENST00000334418.1 |
CCDC84 |
coiled-coil domain containing 84 |
| chr16_+_56995854 | 0.05 |
ENST00000566128.1 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr15_+_44719394 | 0.05 |
ENST00000260327.4 ENST00000396780.1 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr12_+_111843749 | 0.05 |
ENST00000341259.2 |
SH2B3 |
SH2B adaptor protein 3 |
| chr4_-_140223614 | 0.05 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr22_+_20105259 | 0.05 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr3_-_157251383 | 0.05 |
ENST00000487753.1 ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr15_-_57025675 | 0.05 |
ENST00000558320.1 |
ZNF280D |
zinc finger protein 280D |
| chr6_+_10585979 | 0.05 |
ENST00000265012.4 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr11_-_327537 | 0.04 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr5_+_133707252 | 0.04 |
ENST00000506787.1 ENST00000507277.1 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr22_+_20105012 | 0.04 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr18_+_21718924 | 0.04 |
ENST00000399496.3 |
CABYR |
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr7_+_75932863 | 0.04 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr18_+_21719018 | 0.04 |
ENST00000585037.1 ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR |
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr2_-_63815860 | 0.04 |
ENST00000272321.7 ENST00000431065.1 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr17_-_40897043 | 0.04 |
ENST00000428826.2 ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1 |
enhancer of zeste homolog 1 (Drosophila) |
| chr9_+_130186653 | 0.04 |
ENST00000342483.5 ENST00000543471.1 |
ZNF79 |
zinc finger protein 79 |
| chr8_-_131455835 | 0.04 |
ENST00000518721.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr5_-_132166579 | 0.04 |
ENST00000378679.3 |
SHROOM1 |
shroom family member 1 |
| chr19_-_19144243 | 0.04 |
ENST00000594445.1 ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2 |
SURP and G patch domain containing 2 |
| chr1_+_43637996 | 0.04 |
ENST00000528956.1 ENST00000529956.1 |
WDR65 |
WD repeat domain 65 |
| chr1_-_32827682 | 0.04 |
ENST00000432622.1 |
FAM229A |
family with sequence similarity 229, member A |
| chr1_-_24306835 | 0.04 |
ENST00000484146.2 |
SRSF10 |
serine/arginine-rich splicing factor 10 |
| chr10_-_27443155 | 0.04 |
ENST00000427324.1 ENST00000326799.3 |
YME1L1 |
YME1-like 1 ATPase |
| chr1_+_154975110 | 0.04 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr6_-_28973037 | 0.04 |
ENST00000377179.3 |
ZNF311 |
zinc finger protein 311 |
| chr2_-_58468437 | 0.04 |
ENST00000403676.1 ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL |
Fanconi anemia, complementation group L |
| chr9_-_123639445 | 0.04 |
ENST00000312189.6 |
PHF19 |
PHD finger protein 19 |
| chr10_+_35416223 | 0.04 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chrX_+_10031499 | 0.04 |
ENST00000454666.1 |
WWC3 |
WWC family member 3 |
| chr15_+_64386261 | 0.04 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr17_-_43128943 | 0.04 |
ENST00000588499.1 ENST00000593094.1 |
DCAKD |
dephospho-CoA kinase domain containing |
| chr1_+_160051319 | 0.04 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr3_-_136471204 | 0.04 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr22_+_20104947 | 0.04 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
| chr2_+_79252834 | 0.04 |
ENST00000409471.1 |
REG3G |
regenerating islet-derived 3 gamma |
| chr14_-_77737543 | 0.04 |
ENST00000298352.4 |
NGB |
neuroglobin |
| chr16_+_56995762 | 0.04 |
ENST00000200676.3 ENST00000379780.2 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr9_+_130374537 | 0.03 |
ENST00000373302.3 ENST00000373299.1 |
STXBP1 |
syntaxin binding protein 1 |
| chr19_+_37464063 | 0.03 |
ENST00000586353.1 ENST00000433993.1 ENST00000592567.1 |
ZNF568 |
zinc finger protein 568 |
| chr1_-_24306798 | 0.03 |
ENST00000374452.5 ENST00000492112.2 ENST00000343255.5 ENST00000344989.6 |
SRSF10 |
serine/arginine-rich splicing factor 10 |
| chr2_+_232575128 | 0.03 |
ENST00000412128.1 |
PTMA |
prothymosin, alpha |
| chr13_-_113862401 | 0.03 |
ENST00000375459.1 |
PCID2 |
PCI domain containing 2 |
| chr6_+_30103885 | 0.03 |
ENST00000396581.1 |
TRIM40 |
tripartite motif containing 40 |
| chr14_+_36295504 | 0.03 |
ENST00000216807.7 |
BRMS1L |
breast cancer metastasis-suppressor 1-like |
| chr5_+_31532373 | 0.03 |
ENST00000325366.9 ENST00000355907.3 ENST00000507818.2 |
C5orf22 |
chromosome 5 open reading frame 22 |
| chr4_-_106629796 | 0.03 |
ENST00000416543.1 ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12 |
integrator complex subunit 12 |
| chr1_-_155214475 | 0.03 |
ENST00000428024.3 |
GBA |
glucosidase, beta, acid |
| chr4_+_113558272 | 0.03 |
ENST00000509061.1 ENST00000508577.1 ENST00000513553.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr11_-_89224508 | 0.03 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_+_44754126 | 0.03 |
ENST00000449836.1 ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502 |
zinc finger protein 502 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.7 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |