Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for E4F1
Z-value: 1.10
Transcription factors associated with E4F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E4F1
|
ENSG00000167967.11 | E4F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E4F1 | hg19_v2_chr16_+_2273558_2273637 | 0.35 | 3.9e-01 | Click! |
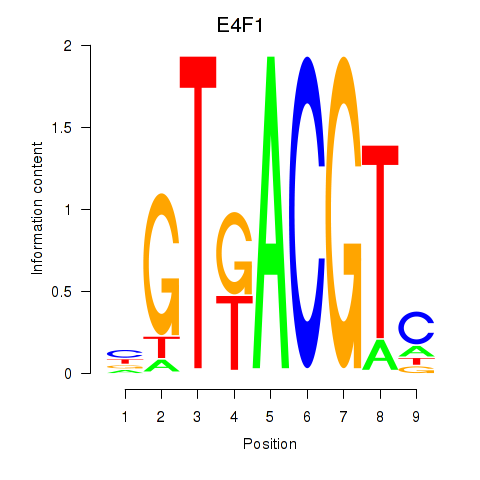
Activity profile of E4F1 motif
Sorted Z-values of E4F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E4F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_58179582 | 1.21 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr1_+_2004901 | 1.03 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr1_+_2005425 | 0.99 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr6_+_28048753 | 0.69 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr17_-_4643114 | 0.63 |
ENST00000293778.6 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
| chr11_+_111411384 | 0.47 |
ENST00000375615.3 ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN |
layilin |
| chr19_+_45504688 | 0.39 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr8_-_95274536 | 0.36 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr7_-_36406750 | 0.34 |
ENST00000453212.1 ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895 |
KIAA0895 |
| chr4_+_75310851 | 0.33 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr17_-_7165662 | 0.33 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr17_+_35294075 | 0.33 |
ENST00000254457.5 |
LHX1 |
LIM homeobox 1 |
| chr8_-_66754172 | 0.32 |
ENST00000401827.3 |
PDE7A |
phosphodiesterase 7A |
| chr7_-_140624499 | 0.32 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr7_+_30174426 | 0.31 |
ENST00000324453.8 |
C7orf41 |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr4_+_75311019 | 0.31 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr18_+_23806437 | 0.28 |
ENST00000578121.1 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr6_+_139456226 | 0.25 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr1_+_26496362 | 0.21 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr14_-_23388338 | 0.21 |
ENST00000555209.1 ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23 |
RNA binding motif protein 23 |
| chr6_-_34664612 | 0.20 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr10_-_50970322 | 0.20 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr5_-_95297534 | 0.19 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr10_-_50970382 | 0.19 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr11_-_77185094 | 0.19 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chrX_+_153237740 | 0.19 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr17_+_41476327 | 0.18 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr4_+_47487285 | 0.16 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr6_+_30029008 | 0.16 |
ENST00000332435.5 ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1 |
zinc ribbon domain containing 1 |
| chr22_+_39101728 | 0.15 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr2_-_64881018 | 0.15 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr9_+_131644388 | 0.14 |
ENST00000372600.4 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr9_+_131644781 | 0.13 |
ENST00000259324.5 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr2_-_220408430 | 0.13 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr2_-_75426826 | 0.13 |
ENST00000305249.5 |
TACR1 |
tachykinin receptor 1 |
| chr9_-_131644202 | 0.13 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr9_+_131644398 | 0.13 |
ENST00000372599.3 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr17_+_18087553 | 0.13 |
ENST00000399138.4 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
| chr22_+_18121356 | 0.12 |
ENST00000317582.5 ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr9_-_131644306 | 0.12 |
ENST00000302586.3 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr3_+_150126101 | 0.12 |
ENST00000361875.3 ENST00000361136.2 |
TSC22D2 |
TSC22 domain family, member 2 |
| chr17_-_36831156 | 0.12 |
ENST00000325814.5 |
C17orf96 |
chromosome 17 open reading frame 96 |
| chr16_+_68057153 | 0.12 |
ENST00000358896.6 ENST00000568099.2 |
DUS2 |
dihydrouridine synthase 2 |
| chr16_+_68056844 | 0.11 |
ENST00000565263.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr6_-_32812420 | 0.11 |
ENST00000374881.2 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr16_+_68057179 | 0.10 |
ENST00000567100.1 ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr7_-_129592471 | 0.10 |
ENST00000473814.2 ENST00000490974.1 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr22_-_41252962 | 0.10 |
ENST00000216218.3 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr5_-_95297678 | 0.10 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr1_+_70820451 | 0.10 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr22_+_18121562 | 0.10 |
ENST00000355028.3 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr17_+_4643300 | 0.09 |
ENST00000433935.1 |
ZMYND15 |
zinc finger, MYND-type containing 15 |
| chr9_-_99382065 | 0.09 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chrX_-_153236819 | 0.09 |
ENST00000354233.3 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chr1_-_8086343 | 0.09 |
ENST00000474874.1 ENST00000469499.1 ENST00000377482.5 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
| chr16_+_29911666 | 0.09 |
ENST00000563177.1 ENST00000483405.1 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
| chr1_-_47184745 | 0.09 |
ENST00000544071.1 |
EFCAB14 |
EF-hand calcium binding domain 14 |
| chr17_+_4643337 | 0.09 |
ENST00000592813.1 |
ZMYND15 |
zinc finger, MYND-type containing 15 |
| chr3_+_184053703 | 0.08 |
ENST00000450976.1 ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A |
family with sequence similarity 131, member A |
| chr14_+_94492674 | 0.08 |
ENST00000203664.5 ENST00000553723.1 |
OTUB2 |
OTU domain, ubiquitin aldehyde binding 2 |
| chr7_-_100026280 | 0.08 |
ENST00000360951.4 ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
| chrX_-_153237258 | 0.08 |
ENST00000310441.7 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chr19_+_52693259 | 0.07 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr10_+_119000604 | 0.07 |
ENST00000298472.5 |
SLC18A2 |
solute carrier family 18 (vesicular monoamine transporter), member 2 |
| chr1_+_180199393 | 0.07 |
ENST00000263726.2 |
LHX4 |
LIM homeobox 4 |
| chr20_-_44420507 | 0.07 |
ENST00000243938.4 |
WFDC3 |
WAP four-disulfide core domain 3 |
| chr3_-_107941209 | 0.07 |
ENST00000492106.1 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr6_-_31774714 | 0.07 |
ENST00000375661.5 |
LSM2 |
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_+_66188475 | 0.07 |
ENST00000311034.2 |
NPAS4 |
neuronal PAS domain protein 4 |
| chr11_-_6633799 | 0.07 |
ENST00000299424.4 |
TAF10 |
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr7_-_129592700 | 0.07 |
ENST00000472396.1 ENST00000355621.3 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr4_-_54930790 | 0.06 |
ENST00000263921.3 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
| chr12_-_93836028 | 0.06 |
ENST00000318066.2 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
| chr6_+_47445467 | 0.06 |
ENST00000359314.5 |
CD2AP |
CD2-associated protein |
| chr2_+_75185619 | 0.06 |
ENST00000483063.1 |
POLE4 |
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr10_+_64893039 | 0.05 |
ENST00000277746.6 ENST00000435510.2 |
NRBF2 |
nuclear receptor binding factor 2 |
| chr16_+_29911864 | 0.05 |
ENST00000308748.5 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
| chr16_-_3767551 | 0.05 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr21_-_40720974 | 0.05 |
ENST00000380748.1 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr12_+_122150646 | 0.05 |
ENST00000449592.2 |
TMEM120B |
transmembrane protein 120B |
| chr3_+_101292939 | 0.05 |
ENST00000265260.3 ENST00000469941.1 ENST00000296024.5 |
PCNP |
PEST proteolytic signal containing nuclear protein |
| chr6_-_53213587 | 0.05 |
ENST00000542638.1 ENST00000370913.5 ENST00000541407.1 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
| chr1_-_32110467 | 0.05 |
ENST00000440872.2 ENST00000373703.4 |
PEF1 |
penta-EF-hand domain containing 1 |
| chr16_-_3767506 | 0.04 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr20_-_44600810 | 0.04 |
ENST00000322927.2 ENST00000426788.1 |
ZNF335 |
zinc finger protein 335 |
| chr1_+_85527987 | 0.04 |
ENST00000326813.8 ENST00000294664.6 ENST00000528899.1 |
WDR63 |
WD repeat domain 63 |
| chr1_+_9599540 | 0.04 |
ENST00000302692.6 |
SLC25A33 |
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr9_+_125027127 | 0.04 |
ENST00000441707.1 ENST00000373723.5 ENST00000373729.1 |
MRRF |
mitochondrial ribosome recycling factor |
| chr11_+_3819049 | 0.04 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr17_+_7211656 | 0.04 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr5_-_133706695 | 0.04 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr14_-_39901618 | 0.04 |
ENST00000554932.1 ENST00000298097.7 |
FBXO33 |
F-box protein 33 |
| chr21_-_40720995 | 0.04 |
ENST00000380749.5 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr2_-_62115659 | 0.04 |
ENST00000544185.1 |
CCT4 |
chaperonin containing TCP1, subunit 4 (delta) |
| chr14_-_50053081 | 0.04 |
ENST00000396020.3 ENST00000245458.6 |
RPS29 |
ribosomal protein S29 |
| chr17_-_80231557 | 0.04 |
ENST00000392334.2 ENST00000314028.6 |
CSNK1D |
casein kinase 1, delta |
| chr19_+_54704610 | 0.04 |
ENST00000302907.4 |
RPS9 |
ribosomal protein S9 |
| chr4_-_111558135 | 0.04 |
ENST00000394598.2 ENST00000394595.3 |
PITX2 |
paired-like homeodomain 2 |
| chr19_+_54704990 | 0.04 |
ENST00000391753.2 |
RPS9 |
ribosomal protein S9 |
| chr1_+_110527308 | 0.03 |
ENST00000369799.5 |
AHCYL1 |
adenosylhomocysteinase-like 1 |
| chr2_-_62115725 | 0.03 |
ENST00000538252.1 ENST00000544079.1 ENST00000394440.3 |
CCT4 |
chaperonin containing TCP1, subunit 4 (delta) |
| chr15_-_53082178 | 0.03 |
ENST00000305901.5 |
ONECUT1 |
one cut homeobox 1 |
| chr9_-_99381660 | 0.03 |
ENST00000375240.3 ENST00000463569.1 |
CDC14B |
cell division cycle 14B |
| chr12_-_93835665 | 0.03 |
ENST00000552442.1 ENST00000550657.1 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
| chr11_-_67276100 | 0.03 |
ENST00000301488.3 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr19_+_1941117 | 0.03 |
ENST00000255641.8 |
CSNK1G2 |
casein kinase 1, gamma 2 |
| chr16_-_27561209 | 0.03 |
ENST00000356183.4 ENST00000561623.1 |
GTF3C1 |
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr17_+_7476136 | 0.03 |
ENST00000582169.1 ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1 |
eukaryotic translation initiation factor 4A1 |
| chr6_-_144416737 | 0.03 |
ENST00000367569.2 |
SF3B5 |
splicing factor 3b, subunit 5, 10kDa |
| chr6_+_32812568 | 0.03 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr17_-_80231300 | 0.03 |
ENST00000398519.5 ENST00000580446.1 |
CSNK1D |
casein kinase 1, delta |
| chr14_+_105219437 | 0.02 |
ENST00000329967.6 ENST00000347067.5 ENST00000553810.1 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
| chr17_+_53342311 | 0.02 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr5_+_68513557 | 0.02 |
ENST00000256441.4 |
MRPS36 |
mitochondrial ribosomal protein S36 |
| chr11_+_66059339 | 0.02 |
ENST00000327259.4 |
TMEM151A |
transmembrane protein 151A |
| chr9_-_125027079 | 0.02 |
ENST00000417201.3 |
RBM18 |
RNA binding motif protein 18 |
| chr17_+_42385927 | 0.02 |
ENST00000426726.3 ENST00000590941.1 ENST00000225441.7 |
RUNDC3A |
RUN domain containing 3A |
| chr6_-_163148780 | 0.02 |
ENST00000366892.1 ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2 |
parkin RBR E3 ubiquitin protein ligase |
| chr9_+_125026882 | 0.02 |
ENST00000297908.3 ENST00000373730.3 ENST00000546115.1 ENST00000344641.3 |
MRRF |
mitochondrial ribosome recycling factor |
| chr1_-_16482554 | 0.02 |
ENST00000358432.5 |
EPHA2 |
EPH receptor A2 |
| chr3_+_185303962 | 0.02 |
ENST00000296257.5 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr4_+_1873100 | 0.02 |
ENST00000508803.1 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
| chr7_-_86849883 | 0.01 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr8_-_29208183 | 0.01 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr3_-_107941230 | 0.01 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr6_+_44355257 | 0.00 |
ENST00000371477.3 |
CDC5L |
cell division cycle 5-like |
| chr3_+_140660634 | 0.00 |
ENST00000446041.2 ENST00000507429.1 ENST00000324194.6 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr17_+_39845134 | 0.00 |
ENST00000591776.1 ENST00000469257.1 |
EIF1 |
eukaryotic translation initiation factor 1 |
| chr22_+_32340447 | 0.00 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr10_-_119134918 | 0.00 |
ENST00000334464.5 |
PDZD8 |
PDZ domain containing 8 |
| chr8_+_94929168 | 0.00 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.3 | GO:0060067 | cervix development(GO:0060067) |
| 0.1 | 0.6 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.6 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |