Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for EBF3
Z-value: 1.22
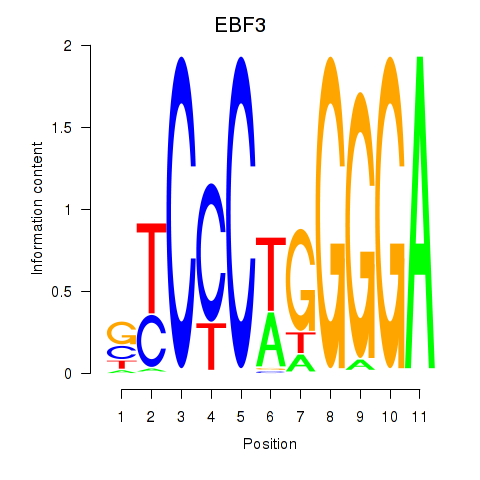
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg19_v2_chr10_-_131762105_131762105 | 0.96 | 1.4e-04 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_68269971 | 1.63 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr16_+_67233007 | 1.40 |
ENST00000360833.1 ENST00000393997.2 |
ELMO3 |
engulfment and cell motility 3 |
| chr1_+_35220613 | 1.31 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr1_-_1051455 | 1.28 |
ENST00000379339.1 ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159 |
chromosome 1 open reading frame 159 |
| chr1_-_242612779 | 1.23 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr1_-_242687989 | 1.19 |
ENST00000442594.2 |
PLD5 |
phospholipase D family, member 5 |
| chr19_-_36004543 | 1.07 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr3_-_189840223 | 1.03 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr1_-_159915386 | 0.96 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr17_-_56494713 | 0.95 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494882 | 0.94 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr16_-_85784718 | 0.94 |
ENST00000602766.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr17_-_56494908 | 0.94 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_39677971 | 0.94 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr16_+_67233412 | 0.91 |
ENST00000477898.1 |
ELMO3 |
engulfment and cell motility 3 |
| chr16_-_85784634 | 0.90 |
ENST00000284245.4 ENST00000602914.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr11_+_1860682 | 0.89 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr16_-_85784557 | 0.89 |
ENST00000602675.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr11_+_1860832 | 0.88 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr1_+_14925173 | 0.87 |
ENST00000376030.2 ENST00000503743.1 ENST00000422387.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr1_+_26869597 | 0.80 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr18_+_34124507 | 0.79 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr11_+_1860200 | 0.79 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr15_+_90728145 | 0.75 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_+_109792641 | 0.74 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr18_-_28681950 | 0.73 |
ENST00000251081.6 |
DSC2 |
desmocollin 2 |
| chr14_+_96671016 | 0.71 |
ENST00000542454.2 ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2 RP11-404P21.8 |
bradykinin receptor B2 Uncharacterized protein |
| chr1_+_153330322 | 0.70 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr1_+_2004901 | 0.67 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr2_-_31361543 | 0.66 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chrX_+_105937068 | 0.64 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_2005425 | 0.63 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr14_-_105635090 | 0.63 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr17_-_7493390 | 0.58 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr2_-_238499303 | 0.57 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr4_+_1795012 | 0.56 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr8_-_139926236 | 0.56 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr13_+_26042960 | 0.53 |
ENST00000255283.8 |
ATP8A2 |
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr1_-_117210290 | 0.52 |
ENST00000369483.1 ENST00000369486.3 |
IGSF3 |
immunoglobulin superfamily, member 3 |
| chr11_+_124933191 | 0.52 |
ENST00000532000.1 ENST00000308074.4 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr1_-_24513737 | 0.51 |
ENST00000374421.3 ENST00000374418.3 ENST00000327535.1 ENST00000327575.2 |
IFNLR1 |
interferon, lambda receptor 1 |
| chr11_+_124932986 | 0.51 |
ENST00000407458.1 ENST00000298280.5 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr11_+_124932955 | 0.51 |
ENST00000403796.2 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr5_-_175964366 | 0.49 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr19_-_38743878 | 0.49 |
ENST00000587515.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr2_-_238499725 | 0.48 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr2_-_238499337 | 0.48 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr2_-_238499131 | 0.47 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr9_-_117111222 | 0.47 |
ENST00000374079.4 |
AKNA |
AT-hook transcription factor |
| chr1_+_3388181 | 0.47 |
ENST00000418137.1 ENST00000413250.2 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chrX_-_33229636 | 0.44 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr14_+_67999999 | 0.42 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr19_+_54369608 | 0.42 |
ENST00000336967.3 |
MYADM |
myeloid-associated differentiation marker |
| chr16_+_4838412 | 0.41 |
ENST00000589327.1 |
SMIM22 |
small integral membrane protein 22 |
| chr8_+_103563792 | 0.41 |
ENST00000285402.3 |
ODF1 |
outer dense fiber of sperm tails 1 |
| chr2_+_220325977 | 0.41 |
ENST00000396686.1 ENST00000396689.2 |
SPEG |
SPEG complex locus |
| chr2_+_220325441 | 0.40 |
ENST00000396688.1 |
SPEG |
SPEG complex locus |
| chr16_+_4838393 | 0.40 |
ENST00000589721.1 |
SMIM22 |
small integral membrane protein 22 |
| chr1_+_33231268 | 0.40 |
ENST00000373480.1 |
KIAA1522 |
KIAA1522 |
| chr11_+_128562372 | 0.38 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_43737939 | 0.37 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr6_-_170599561 | 0.37 |
ENST00000366756.3 |
DLL1 |
delta-like 1 (Drosophila) |
| chr6_+_43738444 | 0.37 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr10_-_73533255 | 0.35 |
ENST00000394957.3 |
C10orf54 |
chromosome 10 open reading frame 54 |
| chr3_-_59035673 | 0.34 |
ENST00000491845.1 ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67 |
chromosome 3 open reading frame 67 |
| chr2_-_75796837 | 0.34 |
ENST00000233712.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chrX_+_38420623 | 0.34 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr6_+_18155560 | 0.34 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr5_-_141257954 | 0.33 |
ENST00000456271.1 ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1 |
protocadherin 1 |
| chr4_+_48988259 | 0.33 |
ENST00000226432.4 |
CWH43 |
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
| chr17_+_7348658 | 0.32 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chrX_+_17393543 | 0.32 |
ENST00000380060.3 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr6_+_18155632 | 0.32 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr8_+_21912328 | 0.32 |
ENST00000432128.1 ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN |
dematin actin binding protein |
| chr12_-_87232644 | 0.31 |
ENST00000549405.2 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_+_22436635 | 0.31 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr10_-_47151341 | 0.31 |
ENST00000422732.2 |
LINC00842 |
long intergenic non-protein coding RNA 842 |
| chr2_+_26568965 | 0.31 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr8_+_22436248 | 0.30 |
ENST00000308354.7 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr17_-_80023401 | 0.30 |
ENST00000354321.7 ENST00000306796.5 |
DUS1L |
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr2_-_64371546 | 0.30 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr17_+_7348374 | 0.30 |
ENST00000306071.2 ENST00000572857.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chrX_-_119445306 | 0.29 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr10_-_105615164 | 0.29 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chrX_-_119445263 | 0.28 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr2_-_46769694 | 0.28 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr4_+_7194247 | 0.28 |
ENST00000507866.2 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
| chr8_-_145641864 | 0.27 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chrX_+_128913906 | 0.27 |
ENST00000356892.3 |
SASH3 |
SAM and SH3 domain containing 3 |
| chr7_+_144052381 | 0.27 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_110577229 | 0.26 |
ENST00000369795.3 ENST00000369794.2 |
STRIP1 |
striatin interacting protein 1 |
| chr22_-_46933067 | 0.26 |
ENST00000262738.3 ENST00000395964.1 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr10_+_99332529 | 0.25 |
ENST00000455090.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chrX_-_152989798 | 0.25 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr7_+_29874341 | 0.24 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chrX_+_48644962 | 0.24 |
ENST00000376670.3 ENST00000376665.3 |
GATA1 |
GATA binding protein 1 (globin transcription factor 1) |
| chrX_-_48827976 | 0.24 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr21_-_45681765 | 0.23 |
ENST00000431166.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr2_+_103378472 | 0.23 |
ENST00000412401.2 |
TMEM182 |
transmembrane protein 182 |
| chr13_-_52980263 | 0.23 |
ENST00000258613.4 ENST00000544466.1 |
THSD1 |
thrombospondin, type I, domain containing 1 |
| chr17_-_73874654 | 0.23 |
ENST00000254816.2 |
TRIM47 |
tripartite motif containing 47 |
| chr4_-_90758227 | 0.23 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr7_+_107110488 | 0.22 |
ENST00000304402.4 |
GPR22 |
G protein-coupled receptor 22 |
| chr11_-_72433346 | 0.22 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_+_41937131 | 0.22 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr9_+_112403088 | 0.22 |
ENST00000448454.2 |
PALM2 |
paralemmin 2 |
| chr1_-_26197744 | 0.22 |
ENST00000374296.3 |
PAQR7 |
progestin and adipoQ receptor family member VII |
| chr12_+_7282795 | 0.21 |
ENST00000266546.6 |
CLSTN3 |
calsyntenin 3 |
| chrX_-_46759138 | 0.21 |
ENST00000377879.3 |
CXorf31 |
chromosome X open reading frame 31 |
| chr17_+_1182948 | 0.21 |
ENST00000333813.3 |
TUSC5 |
tumor suppressor candidate 5 |
| chr19_+_50180317 | 0.21 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr1_+_247579451 | 0.21 |
ENST00000391828.3 ENST00000366497.2 |
NLRP3 |
NLR family, pyrin domain containing 3 |
| chr16_-_68482440 | 0.21 |
ENST00000219334.5 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr2_+_134877740 | 0.21 |
ENST00000409645.1 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr6_+_71123107 | 0.20 |
ENST00000370479.3 ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A |
family with sequence similarity 135, member A |
| chr18_+_10454594 | 0.20 |
ENST00000355285.5 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
| chr10_+_99332198 | 0.20 |
ENST00000307518.5 ENST00000298808.5 ENST00000370655.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr5_+_34915444 | 0.20 |
ENST00000336767.5 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr19_+_15160130 | 0.20 |
ENST00000427043.3 |
CASP14 |
caspase 14, apoptosis-related cysteine peptidase |
| chrX_-_14891150 | 0.20 |
ENST00000452869.1 ENST00000398334.1 ENST00000324138.3 |
FANCB |
Fanconi anemia, complementation group B |
| chr6_-_17987694 | 0.20 |
ENST00000378814.5 ENST00000378843.2 ENST00000378826.2 ENST00000378816.5 ENST00000259711.6 ENST00000502704.1 |
KIF13A |
kinesin family member 13A |
| chr4_-_90758118 | 0.19 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_-_37764128 | 0.19 |
ENST00000302584.4 |
NEUROD2 |
neuronal differentiation 2 |
| chr3_+_140770183 | 0.19 |
ENST00000310546.2 |
SPSB4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr7_+_48128194 | 0.19 |
ENST00000416681.1 ENST00000331803.4 ENST00000432131.1 |
UPP1 |
uridine phosphorylase 1 |
| chr7_+_48128316 | 0.19 |
ENST00000341253.4 |
UPP1 |
uridine phosphorylase 1 |
| chr19_+_2164126 | 0.19 |
ENST00000398665.3 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
| chr17_-_7193711 | 0.19 |
ENST00000571464.1 |
YBX2 |
Y box binding protein 2 |
| chr4_+_87515454 | 0.18 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr8_-_110620284 | 0.18 |
ENST00000529690.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr9_+_112403059 | 0.18 |
ENST00000374531.2 |
PALM2 |
paralemmin 2 |
| chr17_-_1928621 | 0.18 |
ENST00000331238.6 |
RTN4RL1 |
reticulon 4 receptor-like 1 |
| chr22_-_37823468 | 0.18 |
ENST00000402918.2 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr19_+_33865218 | 0.18 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr7_-_100881109 | 0.18 |
ENST00000308344.5 |
CLDN15 |
claudin 15 |
| chr2_+_159825143 | 0.18 |
ENST00000454300.1 ENST00000263635.6 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chrX_-_49056635 | 0.18 |
ENST00000472598.1 ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP |
synaptophysin |
| chr3_+_52813932 | 0.18 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chrX_-_30993201 | 0.18 |
ENST00000288422.2 ENST00000378932.2 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr12_+_112279782 | 0.18 |
ENST00000550735.2 |
MAPKAPK5 |
mitogen-activated protein kinase-activated protein kinase 5 |
| chr7_+_77167376 | 0.18 |
ENST00000435495.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr10_+_70939983 | 0.17 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr1_+_153747746 | 0.17 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr10_+_111985713 | 0.17 |
ENST00000239007.7 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr3_+_143690640 | 0.17 |
ENST00000315691.3 |
C3orf58 |
chromosome 3 open reading frame 58 |
| chr1_+_174769006 | 0.17 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_-_44497024 | 0.16 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_+_14063278 | 0.16 |
ENST00000254337.6 |
DCAF15 |
DDB1 and CUL4 associated factor 15 |
| chr11_+_118978045 | 0.16 |
ENST00000336702.3 |
C2CD2L |
C2CD2-like |
| chr9_-_44402427 | 0.16 |
ENST00000540551.1 |
BX088651.1 |
LOC100126582 protein; Uncharacterized protein |
| chr11_-_72432950 | 0.16 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr8_+_145582217 | 0.16 |
ENST00000530047.1 ENST00000527078.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr11_+_9482551 | 0.16 |
ENST00000438144.2 ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143 |
zinc finger protein 143 |
| chr3_+_193853927 | 0.16 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr8_-_80680078 | 0.16 |
ENST00000337919.5 ENST00000354724.3 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr4_-_57687847 | 0.15 |
ENST00000504762.1 ENST00000248701.4 ENST00000506738.1 |
SPINK2 |
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) |
| chr12_+_50478977 | 0.15 |
ENST00000381513.4 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_-_74023474 | 0.15 |
ENST00000301607.3 |
EVPL |
envoplakin |
| chr17_-_62009702 | 0.15 |
ENST00000006750.3 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
| chr8_-_28747717 | 0.15 |
ENST00000416984.2 |
INTS9 |
integrator complex subunit 9 |
| chr21_-_45682099 | 0.15 |
ENST00000270172.3 ENST00000418993.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr7_-_71912046 | 0.15 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr8_-_28747424 | 0.15 |
ENST00000523436.1 ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9 |
integrator complex subunit 9 |
| chr1_+_100503643 | 0.15 |
ENST00000370152.3 |
HIAT1 |
hippocampus abundant transcript 1 |
| chr15_-_38856836 | 0.15 |
ENST00000450598.2 ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr17_+_1959369 | 0.15 |
ENST00000576444.1 ENST00000322941.3 |
HIC1 |
hypermethylated in cancer 1 |
| chr19_+_13049413 | 0.14 |
ENST00000316448.5 ENST00000588454.1 |
CALR |
calreticulin |
| chr8_+_145582231 | 0.14 |
ENST00000526338.1 ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr8_-_124286735 | 0.14 |
ENST00000395571.3 |
ZHX1 |
zinc fingers and homeoboxes 1 |
| chr17_+_7308172 | 0.14 |
ENST00000575301.1 |
NLGN2 |
neuroligin 2 |
| chr3_+_6902794 | 0.14 |
ENST00000357716.4 ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7 |
glutamate receptor, metabotropic 7 |
| chr1_-_161147275 | 0.14 |
ENST00000319769.5 ENST00000367998.1 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr6_+_7107999 | 0.14 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr4_-_79860506 | 0.14 |
ENST00000295462.3 ENST00000380645.4 ENST00000512733.1 |
PAQR3 |
progestin and adipoQ receptor family member III |
| chr14_+_22966642 | 0.14 |
ENST00000390496.1 |
TRAJ41 |
T cell receptor alpha joining 41 |
| chr15_+_90544532 | 0.14 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chr18_-_40857493 | 0.14 |
ENST00000255224.3 |
SYT4 |
synaptotagmin IV |
| chr17_-_34890732 | 0.14 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr11_-_113345842 | 0.14 |
ENST00000346454.3 |
DRD2 |
dopamine receptor D2 |
| chr2_+_120189422 | 0.13 |
ENST00000306406.4 |
TMEM37 |
transmembrane protein 37 |
| chr1_-_41950342 | 0.13 |
ENST00000372587.4 |
EDN2 |
endothelin 2 |
| chr11_+_124735282 | 0.13 |
ENST00000397801.1 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr6_+_37012607 | 0.13 |
ENST00000423336.1 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr8_+_145582633 | 0.13 |
ENST00000540505.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr11_-_113346111 | 0.13 |
ENST00000362072.3 |
DRD2 |
dopamine receptor D2 |
| chr6_+_158244223 | 0.13 |
ENST00000392185.3 |
SNX9 |
sorting nexin 9 |
| chr7_+_77167343 | 0.13 |
ENST00000433369.2 ENST00000415482.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr22_+_21321531 | 0.13 |
ENST00000405089.1 ENST00000335375.5 |
AIFM3 |
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr2_+_219135115 | 0.13 |
ENST00000248451.3 ENST00000273077.4 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
| chr22_-_48943199 | 0.13 |
ENST00000407505.3 |
CTA-299D3.8 |
Uncharacterized protein |
| chr3_+_46923670 | 0.13 |
ENST00000427125.2 ENST00000430002.2 |
PTH1R |
parathyroid hormone 1 receptor |
| chr17_-_34890759 | 0.13 |
ENST00000431794.3 |
MYO19 |
myosin XIX |
| chr19_+_46010674 | 0.13 |
ENST00000245932.6 ENST00000592139.1 ENST00000590603.1 |
VASP |
vasodilator-stimulated phosphoprotein |
| chr17_-_6616678 | 0.13 |
ENST00000381074.4 ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr1_+_243419306 | 0.12 |
ENST00000355875.4 ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
| chr2_+_27070964 | 0.12 |
ENST00000288699.6 |
DPYSL5 |
dihydropyrimidinase-like 5 |
| chr14_+_75536280 | 0.12 |
ENST00000238686.8 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr8_-_101964265 | 0.12 |
ENST00000395958.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_+_77056134 | 0.12 |
ENST00000528121.1 ENST00000416398.1 |
ZNF503-AS1 |
ZNF503 antisense RNA 1 |
| chrX_-_74376108 | 0.12 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 2.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 1.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.7 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 0.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.6 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 1.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 1.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.5 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 1.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.5 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.4 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.4 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.3 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 3.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0060584 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.4 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0061054 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.2 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.6 | GO:0070977 | bone maturation(GO:0070977) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0035768 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) branch elongation involved in ureteric bud branching(GO:0060681) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.0 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.5 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 2.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 3.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 2.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |