Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for EGR1_EGR4
Z-value: 0.17
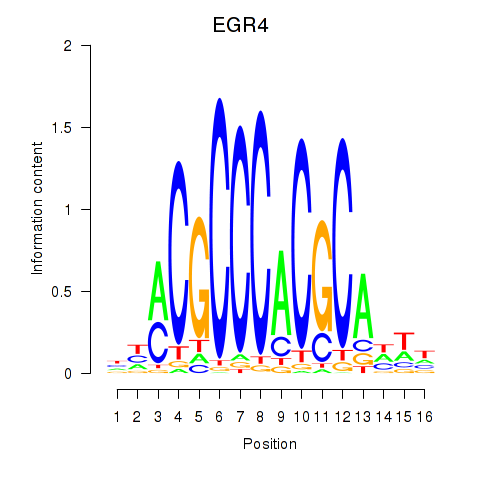
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | EGR1 |
|
EGR4
|
ENSG00000135625.6 | EGR4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR1 | hg19_v2_chr5_+_137801160_137801179 | 0.45 | 2.6e-01 | Click! |
| EGR4 | hg19_v2_chr2_-_73520667_73520833 | 0.19 | 6.5e-01 | Click! |
Activity profile of EGR1_EGR4 motif
Sorted Z-values of EGR1_EGR4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR1_EGR4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44365020 | 0.89 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr2_+_191513959 | 0.35 |
ENST00000337386.5 ENST00000357215.5 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr11_+_124609742 | 0.35 |
ENST00000284292.6 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_124609823 | 0.35 |
ENST00000412681.2 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr16_+_1203194 | 0.34 |
ENST00000348261.5 ENST00000358590.4 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr9_-_124976185 | 0.33 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr19_+_45844032 | 0.32 |
ENST00000589837.1 |
KLC3 |
kinesin light chain 3 |
| chr10_+_49514698 | 0.32 |
ENST00000432379.1 ENST00000429041.1 ENST00000374189.1 |
MAPK8 |
mitogen-activated protein kinase 8 |
| chr1_-_23694794 | 0.31 |
ENST00000374608.3 |
ZNF436 |
zinc finger protein 436 |
| chr1_+_65886244 | 0.30 |
ENST00000344610.8 |
LEPR |
leptin receptor |
| chr17_-_42402138 | 0.30 |
ENST00000592857.1 ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39 |
solute carrier family 25, member 39 |
| chr15_+_72410629 | 0.30 |
ENST00000340912.4 ENST00000544171.1 |
SENP8 |
SUMO/sentrin specific peptidase family member 8 |
| chr7_-_44365216 | 0.29 |
ENST00000358707.3 ENST00000457475.1 ENST00000440254.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr19_-_49371711 | 0.27 |
ENST00000355496.5 ENST00000263265.6 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr19_-_14201776 | 0.27 |
ENST00000269724.5 |
SAMD1 |
sterile alpha motif domain containing 1 |
| chr19_+_47778119 | 0.27 |
ENST00000552360.2 |
PRR24 |
proline rich 24 |
| chr2_+_191513789 | 0.27 |
ENST00000409581.1 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr3_-_38691119 | 0.27 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr1_+_23695680 | 0.26 |
ENST00000454117.1 ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213 |
chromosome 1 open reading frame 213 |
| chr20_-_17539456 | 0.26 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr1_+_203274639 | 0.26 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr17_+_57408994 | 0.26 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr12_+_57482665 | 0.26 |
ENST00000300131.3 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr9_-_130742792 | 0.26 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr14_-_21566731 | 0.25 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr9_-_95896550 | 0.25 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr1_+_6845578 | 0.25 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr1_+_6845497 | 0.25 |
ENST00000473578.1 ENST00000557126.1 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr19_-_46405861 | 0.25 |
ENST00000322217.5 |
MYPOP |
Myb-related transcription factor, partner of profilin |
| chr16_+_67062996 | 0.24 |
ENST00000561924.2 |
CBFB |
core-binding factor, beta subunit |
| chr12_+_77158021 | 0.24 |
ENST00000550876.1 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
| chr12_+_57482877 | 0.23 |
ENST00000342556.6 ENST00000357680.4 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_+_43148059 | 0.23 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr9_-_124976154 | 0.23 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chrX_-_154033793 | 0.23 |
ENST00000369534.3 ENST00000413259.3 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chrX_-_48814278 | 0.22 |
ENST00000455452.1 |
OTUD5 |
OTU domain containing 5 |
| chr18_-_658244 | 0.22 |
ENST00000585033.1 ENST00000323813.3 |
C18orf56 |
chromosome 18 open reading frame 56 |
| chr12_+_70760056 | 0.22 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr20_+_61569463 | 0.22 |
ENST00000266069.3 |
GID8 |
GID complex subunit 8 |
| chr7_+_100797726 | 0.21 |
ENST00000429457.1 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr11_+_63998198 | 0.21 |
ENST00000321460.5 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr2_+_30454390 | 0.21 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr10_+_23728198 | 0.21 |
ENST00000376495.3 |
OTUD1 |
OTU domain containing 1 |
| chr4_-_90758227 | 0.21 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr18_+_3449821 | 0.21 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr11_-_72492903 | 0.21 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr2_+_30370382 | 0.20 |
ENST00000402708.1 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr11_-_72492878 | 0.20 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr19_+_8455200 | 0.20 |
ENST00000601897.1 ENST00000594216.1 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr11_-_62313090 | 0.20 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr4_-_78740511 | 0.20 |
ENST00000504123.1 ENST00000264903.4 ENST00000515441.1 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
| chr11_+_134201911 | 0.19 |
ENST00000389881.3 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr1_-_209979375 | 0.19 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr1_-_209979465 | 0.19 |
ENST00000542854.1 |
IRF6 |
interferon regulatory factor 6 |
| chrX_-_154033661 | 0.19 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr19_+_3572758 | 0.19 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr11_+_67071050 | 0.18 |
ENST00000376757.5 |
SSH3 |
slingshot protein phosphatase 3 |
| chr4_-_90758118 | 0.18 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_67070919 | 0.18 |
ENST00000308127.4 ENST00000308298.7 |
SSH3 |
slingshot protein phosphatase 3 |
| chr8_-_127570603 | 0.18 |
ENST00000304916.3 |
FAM84B |
family with sequence similarity 84, member B |
| chr12_-_53625958 | 0.18 |
ENST00000327550.3 ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG |
retinoic acid receptor, gamma |
| chr9_-_77643189 | 0.18 |
ENST00000376837.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr1_+_65886326 | 0.18 |
ENST00000371059.3 ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR |
leptin receptor |
| chr20_-_22565101 | 0.18 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chrX_-_3631635 | 0.17 |
ENST00000262848.5 |
PRKX |
protein kinase, X-linked |
| chr17_+_37783170 | 0.17 |
ENST00000254079.4 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_+_155051305 | 0.17 |
ENST00000368408.3 |
EFNA3 |
ephrin-A3 |
| chr2_+_23608064 | 0.17 |
ENST00000486442.1 |
KLHL29 |
kelch-like family member 29 |
| chr18_+_3448455 | 0.17 |
ENST00000549780.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr12_-_49351148 | 0.17 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr12_+_54378923 | 0.16 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr19_-_14201507 | 0.16 |
ENST00000533683.2 |
SAMD1 |
sterile alpha motif domain containing 1 |
| chr19_+_7985880 | 0.16 |
ENST00000597584.1 |
SNAPC2 |
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr11_-_46142948 | 0.16 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr12_-_49351228 | 0.16 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr2_-_27718052 | 0.16 |
ENST00000264703.3 |
FNDC4 |
fibronectin type III domain containing 4 |
| chr2_+_220306745 | 0.16 |
ENST00000431523.1 ENST00000396698.1 ENST00000396695.2 |
SPEG |
SPEG complex locus |
| chr7_-_75368248 | 0.16 |
ENST00000434438.2 ENST00000336926.6 |
HIP1 |
huntingtin interacting protein 1 |
| chr22_-_29137771 | 0.15 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr16_+_30194916 | 0.15 |
ENST00000570045.1 ENST00000565497.1 ENST00000570244.1 |
CORO1A |
coronin, actin binding protein, 1A |
| chr6_+_33172407 | 0.15 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_176924562 | 0.15 |
ENST00000359895.2 ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr16_+_2933229 | 0.15 |
ENST00000573965.1 ENST00000572006.1 |
FLYWCH2 |
FLYWCH family member 2 |
| chr2_-_128145498 | 0.15 |
ENST00000409179.2 |
MAP3K2 |
mitogen-activated protein kinase kinase kinase 2 |
| chr17_-_19281203 | 0.15 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr3_+_39093481 | 0.15 |
ENST00000302313.5 ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48 |
WD repeat domain 48 |
| chr7_+_100860949 | 0.15 |
ENST00000305105.2 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
| chr5_+_172483347 | 0.15 |
ENST00000522692.1 ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF |
CREB3 regulatory factor |
| chr21_+_17102311 | 0.15 |
ENST00000285679.6 ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25 |
ubiquitin specific peptidase 25 |
| chr19_+_45844018 | 0.15 |
ENST00000585434.1 |
KLC3 |
kinesin light chain 3 |
| chr15_+_40733387 | 0.15 |
ENST00000416165.1 |
BAHD1 |
bromo adjacent homology domain containing 1 |
| chr1_-_38471156 | 0.15 |
ENST00000373016.3 |
FHL3 |
four and a half LIM domains 3 |
| chr14_+_100531615 | 0.15 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr11_-_18034549 | 0.15 |
ENST00000528200.1 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr19_-_33793430 | 0.14 |
ENST00000498907.2 |
CEBPA |
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr19_+_45843994 | 0.14 |
ENST00000391946.2 |
KLC3 |
kinesin light chain 3 |
| chr5_+_139028510 | 0.14 |
ENST00000502336.1 ENST00000520967.1 ENST00000511048.1 |
CXXC5 |
CXXC finger protein 5 |
| chrX_-_48901012 | 0.14 |
ENST00000315869.7 |
TFE3 |
transcription factor binding to IGHM enhancer 3 |
| chr10_-_127511790 | 0.14 |
ENST00000368797.4 ENST00000420761.1 |
UROS |
uroporphyrinogen III synthase |
| chr12_-_58240470 | 0.14 |
ENST00000548823.1 ENST00000398073.2 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_+_43148625 | 0.14 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr1_+_211432700 | 0.14 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr20_+_17207665 | 0.14 |
ENST00000536609.1 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr16_+_3014217 | 0.14 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr10_+_104178946 | 0.13 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr14_+_96342729 | 0.13 |
ENST00000504119.1 |
LINC00617 |
long intergenic non-protein coding RNA 617 |
| chr3_-_88108192 | 0.13 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr22_-_30722912 | 0.13 |
ENST00000215790.7 |
TBC1D10A |
TBC1 domain family, member 10A |
| chr17_-_73178599 | 0.13 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr6_-_53530474 | 0.13 |
ENST00000370905.3 |
KLHL31 |
kelch-like family member 31 |
| chr16_-_67969888 | 0.13 |
ENST00000574576.2 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr12_+_26348582 | 0.13 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr19_-_55691377 | 0.13 |
ENST00000589172.1 |
SYT5 |
synaptotagmin V |
| chr11_+_63998006 | 0.13 |
ENST00000355040.4 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr9_+_137218362 | 0.13 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr1_+_24104869 | 0.13 |
ENST00000246151.4 |
PITHD1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr8_-_75233563 | 0.13 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr4_-_82136114 | 0.13 |
ENST00000395578.1 ENST00000418486.2 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
| chr11_-_18034701 | 0.13 |
ENST00000265965.5 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr19_+_10400615 | 0.13 |
ENST00000221980.4 |
ICAM5 |
intercellular adhesion molecule 5, telencephalin |
| chr11_-_46142615 | 0.13 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr1_-_6321035 | 0.13 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr7_+_100797678 | 0.13 |
ENST00000337619.5 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr17_+_17380294 | 0.13 |
ENST00000268711.3 ENST00000580462.1 |
MED9 |
mediator complex subunit 9 |
| chr3_+_37903432 | 0.12 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr5_+_179921430 | 0.12 |
ENST00000393356.1 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr9_-_130661916 | 0.12 |
ENST00000373142.1 ENST00000373146.1 ENST00000373144.3 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr17_-_61920280 | 0.12 |
ENST00000448276.2 ENST00000577990.1 |
SMARCD2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr4_+_38869298 | 0.12 |
ENST00000510213.1 ENST00000515037.1 |
FAM114A1 |
family with sequence similarity 114, member A1 |
| chr20_-_48099182 | 0.12 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr1_+_38273988 | 0.12 |
ENST00000446260.2 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr2_+_232651124 | 0.12 |
ENST00000350033.3 ENST00000412591.1 ENST00000410017.1 ENST00000373608.3 |
COPS7B |
COP9 signalosome subunit 7B |
| chr2_+_46524537 | 0.12 |
ENST00000263734.3 |
EPAS1 |
endothelial PAS domain protein 1 |
| chr10_+_131265443 | 0.12 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr19_-_2050852 | 0.12 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr6_-_19804973 | 0.12 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr22_-_30722866 | 0.12 |
ENST00000403477.3 |
TBC1D10A |
TBC1 domain family, member 10A |
| chr21_+_38445539 | 0.12 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr1_+_26146397 | 0.12 |
ENST00000374303.2 ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr9_-_140196703 | 0.12 |
ENST00000356628.2 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
| chr11_-_18034430 | 0.12 |
ENST00000530613.1 ENST00000532389.1 ENST00000529728.1 ENST00000532265.1 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr16_+_3014269 | 0.12 |
ENST00000575885.1 ENST00000571007.1 ENST00000319500.6 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr19_+_46144884 | 0.11 |
ENST00000593161.1 |
AC006132.1 |
chromosome 19 open reading frame 83 |
| chr19_-_35992780 | 0.11 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chrX_-_107334750 | 0.11 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr7_+_43803790 | 0.11 |
ENST00000424330.1 |
BLVRA |
biliverdin reductase A |
| chrX_-_107334790 | 0.11 |
ENST00000217958.3 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr12_-_124457257 | 0.11 |
ENST00000545891.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr1_+_38273818 | 0.11 |
ENST00000373042.4 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr16_+_67063262 | 0.11 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr21_+_27543175 | 0.11 |
ENST00000608591.1 ENST00000609365.1 |
AP000230.1 |
AP000230.1 |
| chr12_+_51632638 | 0.11 |
ENST00000549732.2 |
DAZAP2 |
DAZ associated protein 2 |
| chr9_+_90112117 | 0.11 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr3_-_180707466 | 0.11 |
ENST00000491873.1 ENST00000486355.1 ENST00000382564.2 |
DNAJC19 |
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr9_+_34989638 | 0.11 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr16_+_85646891 | 0.11 |
ENST00000393243.1 |
GSE1 |
Gse1 coiled-coil protein |
| chr19_-_13068012 | 0.11 |
ENST00000316939.1 |
GADD45GIP1 |
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr2_-_25896380 | 0.11 |
ENST00000545439.1 ENST00000407186.1 ENST00000406818.3 ENST00000404103.3 ENST00000407661.3 ENST00000407038.3 ENST00000405222.1 ENST00000288642.8 |
DTNB |
dystrobrevin, beta |
| chr17_+_4613776 | 0.11 |
ENST00000269260.2 |
ARRB2 |
arrestin, beta 2 |
| chr14_-_23822080 | 0.11 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr6_+_35995552 | 0.11 |
ENST00000468133.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr3_-_185542817 | 0.11 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr9_+_841690 | 0.11 |
ENST00000382276.3 |
DMRT1 |
doublesex and mab-3 related transcription factor 1 |
| chr11_-_75062730 | 0.11 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr8_-_102217796 | 0.11 |
ENST00000519744.1 ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706 |
zinc finger protein 706 |
| chr6_-_4135825 | 0.11 |
ENST00000380118.3 ENST00000413766.2 ENST00000361538.2 |
ECI2 |
enoyl-CoA delta isomerase 2 |
| chr14_-_89883412 | 0.11 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr17_-_6947225 | 0.11 |
ENST00000574600.1 ENST00000308009.1 ENST00000447225.1 |
SLC16A11 |
solute carrier family 16, member 11 |
| chr1_+_26147319 | 0.11 |
ENST00000374300.3 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr9_+_127020202 | 0.10 |
ENST00000373600.3 ENST00000320246.5 |
NEK6 |
NIMA-related kinase 6 |
| chr11_-_65640198 | 0.10 |
ENST00000528176.1 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
| chrX_-_51239425 | 0.10 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr5_-_176923803 | 0.10 |
ENST00000506161.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr17_-_8027402 | 0.10 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr7_+_150782945 | 0.10 |
ENST00000463381.1 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr18_+_22006580 | 0.10 |
ENST00000284202.4 |
IMPACT |
impact RWD domain protein |
| chr7_+_127228399 | 0.10 |
ENST00000000233.5 ENST00000415666.1 |
ARF5 |
ADP-ribosylation factor 5 |
| chr17_+_11924129 | 0.10 |
ENST00000353533.5 ENST00000415385.3 |
MAP2K4 |
mitogen-activated protein kinase kinase 4 |
| chr16_+_1359138 | 0.10 |
ENST00000325437.5 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chr5_+_172068232 | 0.10 |
ENST00000520919.1 ENST00000522853.1 ENST00000369800.5 |
NEURL1B |
neuralized E3 ubiquitin protein ligase 1B |
| chr1_-_146644036 | 0.10 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr1_+_235490659 | 0.10 |
ENST00000488594.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chrX_+_134555863 | 0.10 |
ENST00000417443.2 |
LINC00086 |
long intergenic non-protein coding RNA 86 |
| chr8_-_30515693 | 0.10 |
ENST00000355904.4 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr10_-_99393208 | 0.10 |
ENST00000307450.6 |
MORN4 |
MORN repeat containing 4 |
| chrX_+_153237740 | 0.10 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr19_+_39897453 | 0.10 |
ENST00000597629.1 ENST00000248673.3 ENST00000594045.1 ENST00000594442.1 |
ZFP36 |
ZFP36 ring finger protein |
| chr9_+_35829208 | 0.10 |
ENST00000439587.2 ENST00000377991.4 |
TMEM8B |
transmembrane protein 8B |
| chr12_+_50017184 | 0.10 |
ENST00000548825.2 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr7_+_128502871 | 0.10 |
ENST00000249289.4 |
ATP6V1F |
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr15_+_57884086 | 0.10 |
ENST00000380569.2 ENST00000380561.2 ENST00000574161.1 ENST00000572390.1 ENST00000396180.1 ENST00000380560.2 |
GCOM1 |
GRINL1A complex locus 1 |
| chr9_-_86432547 | 0.10 |
ENST00000376365.3 ENST00000376371.2 |
GKAP1 |
G kinase anchoring protein 1 |
| chr10_-_134145321 | 0.10 |
ENST00000368625.4 ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C |
serine/threonine kinase 32C |
| chr14_-_99947121 | 0.10 |
ENST00000329331.3 ENST00000436070.2 |
SETD3 |
SET domain containing 3 |
| chr1_+_212458834 | 0.10 |
ENST00000261461.2 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
| chr9_+_127020503 | 0.10 |
ENST00000545174.1 ENST00000444973.1 ENST00000454453.1 |
NEK6 |
NIMA-related kinase 6 |
| chr2_+_27651519 | 0.10 |
ENST00000379863.3 |
NRBP1 |
nuclear receptor binding protein 1 |
| chr10_-_105452917 | 0.10 |
ENST00000427662.2 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr14_-_51561784 | 0.10 |
ENST00000360392.4 |
TRIM9 |
tripartite motif containing 9 |
| chr6_+_72596604 | 0.10 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.4 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 1.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.1 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0060129 | mesendoderm development(GO:0048382) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.0 | 0.1 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.6 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0052844 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |