Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for ELF3_EHF
Z-value: 1.78
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
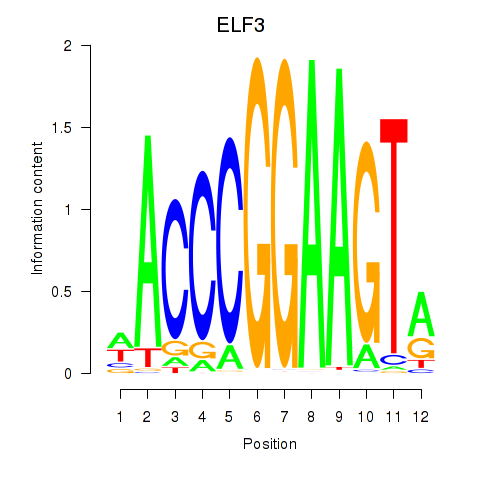
ELF3
|
ENSG00000163435.11 | ELF3 |
|
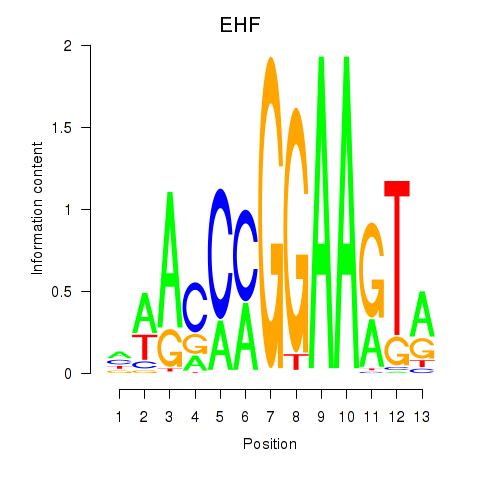
EHF
|
ENSG00000135373.8 | EHF |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EHF | hg19_v2_chr11_+_34642656_34642682 | 0.91 | 1.7e-03 | Click! |
| ELF3 | hg19_v2_chr1_+_201979645_201979721 | 0.66 | 7.6e-02 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_11779174 | 2.55 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr6_-_11779403 | 2.47 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr6_-_11779014 | 2.46 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr15_+_43885252 | 1.73 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr11_-_118122996 | 1.68 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr16_+_67233412 | 1.66 |
ENST00000477898.1 |
ELMO3 |
engulfment and cell motility 3 |
| chr16_+_67233007 | 1.66 |
ENST00000360833.1 ENST00000393997.2 |
ELMO3 |
engulfment and cell motility 3 |
| chr15_+_43985725 | 1.65 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_90744533 | 1.62 |
ENST00000411539.2 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr11_-_119993734 | 1.59 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_119993979 | 1.59 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr19_+_35739897 | 1.48 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr15_+_43985084 | 1.47 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr10_-_98031310 | 1.24 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr19_+_35739782 | 1.20 |
ENST00000347609.4 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr2_-_238499337 | 1.18 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_-_123201337 | 1.17 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr12_+_113344811 | 1.14 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_161056802 | 1.13 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr12_+_113344755 | 1.12 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_161056762 | 1.12 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr19_+_7701985 | 1.04 |
ENST00000595950.1 ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2 |
syntaxin binding protein 2 |
| chr10_-_98031265 | 1.03 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr6_-_11779840 | 1.03 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr7_-_143105941 | 1.02 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr15_+_80987617 | 1.02 |
ENST00000258884.4 ENST00000558464.1 |
ABHD17C |
abhydrolase domain containing 17C |
| chr3_-_57113314 | 0.99 |
ENST00000338458.4 ENST00000468727.1 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr12_-_123187890 | 0.99 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr17_+_48610074 | 0.98 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr8_+_56792377 | 0.97 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr8_+_56792355 | 0.94 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr20_+_1875110 | 0.90 |
ENST00000400068.3 |
SIRPA |
signal-regulatory protein alpha |
| chr1_+_95286151 | 0.90 |
ENST00000467909.1 ENST00000422520.2 ENST00000532427.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr2_-_216003127 | 0.89 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr12_-_6740802 | 0.88 |
ENST00000431922.1 |
LPAR5 |
lysophosphatidic acid receptor 5 |
| chr4_-_84255935 | 0.87 |
ENST00000513463.1 |
HPSE |
heparanase |
| chr4_-_84256024 | 0.85 |
ENST00000311412.5 |
HPSE |
heparanase |
| chr1_+_15250596 | 0.85 |
ENST00000361144.5 |
KAZN |
kazrin, periplakin interacting protein |
| chr11_-_108408895 | 0.84 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chr1_+_31883048 | 0.78 |
ENST00000536859.1 |
SERINC2 |
serine incorporator 2 |
| chr18_-_52989217 | 0.78 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr19_+_6464243 | 0.75 |
ENST00000600229.1 ENST00000356762.3 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr6_-_116381918 | 0.75 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr1_+_86889769 | 0.74 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr3_-_48471454 | 0.73 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr2_-_75788038 | 0.72 |
ENST00000393913.3 ENST00000410113.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr19_+_6464502 | 0.71 |
ENST00000308243.7 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr16_+_4845379 | 0.71 |
ENST00000588606.1 ENST00000586005.1 |
SMIM22 |
small integral membrane protein 22 |
| chr8_+_86089460 | 0.68 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr2_-_238499725 | 0.68 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_-_67072714 | 0.68 |
ENST00000545666.1 ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr18_-_52989525 | 0.67 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr11_-_58345569 | 0.67 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr6_-_143832820 | 0.66 |
ENST00000002165.6 |
FUCA2 |
fucosidase, alpha-L- 2, plasma |
| chr12_-_53343602 | 0.66 |
ENST00000546897.1 ENST00000552551.1 |
KRT8 |
keratin 8 |
| chr1_+_95285896 | 0.66 |
ENST00000446120.2 ENST00000271227.6 ENST00000527077.1 ENST00000529450.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr21_+_43919710 | 0.65 |
ENST00000398341.3 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr19_-_1174226 | 0.63 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr1_-_160990886 | 0.62 |
ENST00000537746.1 |
F11R |
F11 receptor |
| chr8_-_80993010 | 0.61 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr9_+_71944241 | 0.61 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr8_-_134309823 | 0.61 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr8_-_134309335 | 0.61 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr12_+_113344582 | 0.60 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr6_-_42419649 | 0.60 |
ENST00000372922.4 ENST00000541110.1 ENST00000372917.4 |
TRERF1 |
transcriptional regulating factor 1 |
| chr11_+_63753883 | 0.59 |
ENST00000538426.1 ENST00000543004.1 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
| chr19_-_16045220 | 0.57 |
ENST00000326742.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr14_+_61788429 | 0.56 |
ENST00000332981.5 |
PRKCH |
protein kinase C, eta |
| chr19_+_35739280 | 0.55 |
ENST00000602122.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr1_-_27286897 | 0.55 |
ENST00000320567.5 |
C1orf172 |
chromosome 1 open reading frame 172 |
| chr8_-_8243968 | 0.54 |
ENST00000520004.1 |
SGK223 |
Tyrosine-protein kinase SgK223 |
| chr8_-_86253888 | 0.53 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr19_-_56632592 | 0.53 |
ENST00000587279.1 ENST00000270459.3 |
ZNF787 |
zinc finger protein 787 |
| chr2_+_163200848 | 0.52 |
ENST00000233612.4 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr15_+_45422131 | 0.52 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr12_-_50616382 | 0.51 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr12_-_50616122 | 0.50 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr6_-_143832793 | 0.50 |
ENST00000438118.2 |
FUCA2 |
fucosidase, alpha-L- 2, plasma |
| chr2_+_163200598 | 0.50 |
ENST00000437150.2 ENST00000453113.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr2_+_37458928 | 0.49 |
ENST00000439218.1 ENST00000432075.1 |
NDUFAF7 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr1_+_44435646 | 0.49 |
ENST00000255108.3 ENST00000412950.2 ENST00000396758.2 |
DPH2 |
DPH2 homolog (S. cerevisiae) |
| chr18_-_28742813 | 0.49 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr6_+_109761898 | 0.46 |
ENST00000258052.3 |
SMPD2 |
sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase) |
| chr6_+_30848557 | 0.46 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_64885111 | 0.45 |
ENST00000528598.1 ENST00000310597.4 |
ZNHIT2 |
zinc finger, HIT-type containing 2 |
| chrX_+_82763265 | 0.45 |
ENST00000373200.2 |
POU3F4 |
POU class 3 homeobox 4 |
| chr17_-_38256973 | 0.45 |
ENST00000246672.3 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
| chr16_-_402639 | 0.45 |
ENST00000262320.3 |
AXIN1 |
axin 1 |
| chr16_-_11680791 | 0.44 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr1_+_205473720 | 0.44 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr15_+_77287426 | 0.44 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr19_-_16045619 | 0.43 |
ENST00000402119.4 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr3_-_47324242 | 0.43 |
ENST00000456548.1 ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9 |
kinesin family member 9 |
| chr19_-_16045665 | 0.43 |
ENST00000248041.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr11_+_35211429 | 0.43 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr18_+_55816546 | 0.43 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_96931834 | 0.42 |
ENST00000488633.1 |
CIAO1 |
cytosolic iron-sulfur protein assembly 1 |
| chr9_-_117150243 | 0.42 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr16_+_57673430 | 0.42 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr5_+_149737202 | 0.42 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr1_-_183560011 | 0.42 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr1_+_32042105 | 0.41 |
ENST00000457433.2 ENST00000441210.2 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr4_-_89152474 | 0.41 |
ENST00000515655.1 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr13_-_99852916 | 0.40 |
ENST00000426037.2 ENST00000445737.2 |
UBAC2-AS1 |
UBAC2 antisense RNA 1 |
| chr3_+_119217422 | 0.40 |
ENST00000466984.1 |
TIMMDC1 |
translocase of inner mitochondrial membrane domain containing 1 |
| chr1_+_32042131 | 0.40 |
ENST00000271064.7 ENST00000537531.1 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr1_-_28520447 | 0.40 |
ENST00000539896.1 |
PTAFR |
platelet-activating factor receptor |
| chr19_-_54676846 | 0.40 |
ENST00000301187.4 |
TMC4 |
transmembrane channel-like 4 |
| chr4_+_166128735 | 0.39 |
ENST00000226725.6 |
KLHL2 |
kelch-like family member 2 |
| chr17_-_76123101 | 0.39 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
| chr21_+_43619796 | 0.39 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr12_-_64616019 | 0.39 |
ENST00000311915.8 ENST00000398055.3 ENST00000544871.1 |
C12orf66 |
chromosome 12 open reading frame 66 |
| chr1_+_24117627 | 0.39 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr6_+_53659746 | 0.38 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr19_-_55660561 | 0.38 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr11_-_9336234 | 0.38 |
ENST00000528080.1 |
TMEM41B |
transmembrane protein 41B |
| chr10_-_103880209 | 0.38 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr19_+_1077393 | 0.37 |
ENST00000590577.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr16_+_84682108 | 0.37 |
ENST00000564996.1 ENST00000258157.5 ENST00000567410.1 |
KLHL36 |
kelch-like family member 36 |
| chr11_+_70244510 | 0.37 |
ENST00000346329.3 ENST00000301843.8 ENST00000376561.3 |
CTTN |
cortactin |
| chr1_-_200379129 | 0.37 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr11_+_66824276 | 0.37 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr1_-_153029980 | 0.36 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr1_+_18081804 | 0.36 |
ENST00000375406.1 |
ACTL8 |
actin-like 8 |
| chr2_+_106361333 | 0.36 |
ENST00000233154.4 ENST00000451463.2 |
NCK2 |
NCK adaptor protein 2 |
| chr5_-_35230434 | 0.35 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr18_-_67873078 | 0.35 |
ENST00000255674.6 |
RTTN |
rotatin |
| chr1_+_154947126 | 0.35 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr11_+_66824346 | 0.34 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr1_+_156698234 | 0.34 |
ENST00000368218.4 ENST00000368216.4 |
RRNAD1 |
ribosomal RNA adenine dimethylase domain containing 1 |
| chr14_+_69865401 | 0.34 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr2_+_27665232 | 0.34 |
ENST00000543753.1 ENST00000288873.3 |
KRTCAP3 |
keratinocyte associated protein 3 |
| chr5_+_68711209 | 0.34 |
ENST00000512803.1 |
MARVELD2 |
MARVEL domain containing 2 |
| chr8_+_21911054 | 0.34 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr3_+_119217376 | 0.34 |
ENST00000494664.1 ENST00000493694.1 |
TIMMDC1 |
translocase of inner mitochondrial membrane domain containing 1 |
| chr15_-_83378611 | 0.33 |
ENST00000542200.1 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
| chr12_+_57984965 | 0.33 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr8_+_86089619 | 0.33 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr3_+_66271410 | 0.33 |
ENST00000336733.6 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr2_+_27665289 | 0.33 |
ENST00000407293.1 |
KRTCAP3 |
keratinocyte associated protein 3 |
| chr15_+_75074385 | 0.33 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr9_-_115095883 | 0.32 |
ENST00000450374.1 ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr16_+_15068916 | 0.32 |
ENST00000455313.2 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
| chr14_+_24641062 | 0.32 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr1_-_200379180 | 0.32 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr17_+_41561317 | 0.32 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr6_-_99395787 | 0.32 |
ENST00000369244.2 ENST00000229971.1 |
FBXL4 |
F-box and leucine-rich repeat protein 4 |
| chr1_-_52870059 | 0.32 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr2_+_169659121 | 0.32 |
ENST00000397206.2 ENST00000397209.2 ENST00000421711.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chrX_+_138612889 | 0.32 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr19_-_46234119 | 0.32 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr18_-_67872891 | 0.32 |
ENST00000454359.1 ENST00000437017.1 |
RTTN |
rotatin |
| chr17_+_32612687 | 0.31 |
ENST00000305869.3 |
CCL11 |
chemokine (C-C motif) ligand 11 |
| chr1_+_53662101 | 0.31 |
ENST00000371486.3 |
CPT2 |
carnitine palmitoyltransferase 2 |
| chr11_+_112047087 | 0.31 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr11_+_6226782 | 0.31 |
ENST00000316375.2 |
C11orf42 |
chromosome 11 open reading frame 42 |
| chr17_+_34958001 | 0.31 |
ENST00000250156.7 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr19_-_55791431 | 0.31 |
ENST00000593263.1 ENST00000376343.3 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_+_111303218 | 0.31 |
ENST00000441448.2 |
RPF2 |
ribosome production factor 2 homolog (S. cerevisiae) |
| chr17_-_7165662 | 0.31 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr8_-_130799134 | 0.31 |
ENST00000276708.4 |
GSDMC |
gasdermin C |
| chr3_+_66271489 | 0.31 |
ENST00000536651.1 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr19_-_51466681 | 0.31 |
ENST00000456750.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr18_+_55888767 | 0.31 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr17_+_40714092 | 0.30 |
ENST00000420359.1 ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY |
CoA synthase |
| chr3_-_53080047 | 0.30 |
ENST00000482396.1 ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1 |
Scm-like with four mbt domains 1 |
| chr11_+_35211511 | 0.30 |
ENST00000524922.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr3_-_47324008 | 0.30 |
ENST00000425853.1 |
KIF9 |
kinesin family member 9 |
| chr18_-_33077556 | 0.30 |
ENST00000589273.1 ENST00000586489.1 |
INO80C |
INO80 complex subunit C |
| chr3_-_47324060 | 0.30 |
ENST00000452770.2 |
KIF9 |
kinesin family member 9 |
| chr3_-_79816965 | 0.30 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr11_-_72463421 | 0.30 |
ENST00000393609.3 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_+_24117693 | 0.30 |
ENST00000374503.3 ENST00000374502.3 |
LYPLA2 |
lysophospholipase II |
| chr10_+_95326416 | 0.29 |
ENST00000371481.4 ENST00000371483.4 ENST00000604414.1 |
FFAR4 |
free fatty acid receptor 4 |
| chr6_+_111580508 | 0.29 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chr15_+_43663257 | 0.29 |
ENST00000260383.7 ENST00000564079.1 |
TUBGCP4 |
tubulin, gamma complex associated protein 4 |
| chr1_-_52499443 | 0.29 |
ENST00000371614.1 |
KTI12 |
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr8_-_131028869 | 0.29 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr22_+_50628999 | 0.29 |
ENST00000395827.1 |
TRABD |
TraB domain containing |
| chr11_-_7818520 | 0.29 |
ENST00000329434.2 |
OR5P2 |
olfactory receptor, family 5, subfamily P, member 2 |
| chr4_+_41937131 | 0.29 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr6_+_18155632 | 0.28 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr2_+_169658928 | 0.28 |
ENST00000317647.7 ENST00000445023.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chr19_-_51530916 | 0.28 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr10_-_118765081 | 0.28 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr19_+_19496728 | 0.28 |
ENST00000537887.1 ENST00000417582.2 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr15_-_101835110 | 0.28 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr3_+_169755715 | 0.28 |
ENST00000355897.5 |
GPR160 |
G protein-coupled receptor 160 |
| chr5_+_40841276 | 0.28 |
ENST00000254691.5 |
CARD6 |
caspase recruitment domain family, member 6 |
| chr15_+_74833518 | 0.27 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_-_48470838 | 0.27 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr1_+_153750622 | 0.27 |
ENST00000532853.1 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr1_-_156698181 | 0.27 |
ENST00000313146.6 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr11_-_75236867 | 0.27 |
ENST00000376282.3 ENST00000336898.3 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr1_-_153066998 | 0.27 |
ENST00000368750.3 |
SPRR2E |
small proline-rich protein 2E |
| chr11_+_34642656 | 0.27 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr1_-_52870104 | 0.27 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr11_+_2421718 | 0.27 |
ENST00000380996.5 ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4 |
tumor suppressing subtransferable candidate 4 |
| chr20_-_48532019 | 0.27 |
ENST00000289431.5 |
SPATA2 |
spermatogenesis associated 2 |
| chr14_-_24036943 | 0.26 |
ENST00000556843.1 ENST00000397120.3 ENST00000557189.1 |
AP1G2 |
adaptor-related protein complex 1, gamma 2 subunit |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.6 | 1.9 | GO:0070666 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.4 | 1.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.4 | 1.4 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.3 | 1.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 1.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 | 3.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 2.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 1.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 0.6 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 0.8 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.4 | GO:0043132 | coenzyme A transport(GO:0015880) FAD transport(GO:0015883) coenzyme A transmembrane transport(GO:0035349) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.4 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.4 | GO:0071258 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 | 1.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.6 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.6 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.3 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.3 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 0.4 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 1.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 1.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.5 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.3 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.3 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.4 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.2 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:0009205 | ribonucleoside triphosphate metabolic process(GO:0009199) purine ribonucleoside triphosphate metabolic process(GO:0009205) |
| 0.0 | 0.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.6 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.0 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.8 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 1.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 1.2 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.2 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 2.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 3.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 2.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.2 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0021942 | layer formation in cerebral cortex(GO:0021819) radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) cellular polysaccharide biosynthetic process(GO:0033692) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.7 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.3 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 1.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 3.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 1.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.5 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 3.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 2.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 2.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 2.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.7 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.4 | 1.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.3 | 1.3 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.3 | 2.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.6 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 2.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.6 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.4 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.1 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 1.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) ubiquinone binding(GO:0048039) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 3.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 3.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 4.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |