Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for EN1_ESX1_GBX1
Z-value: 0.45
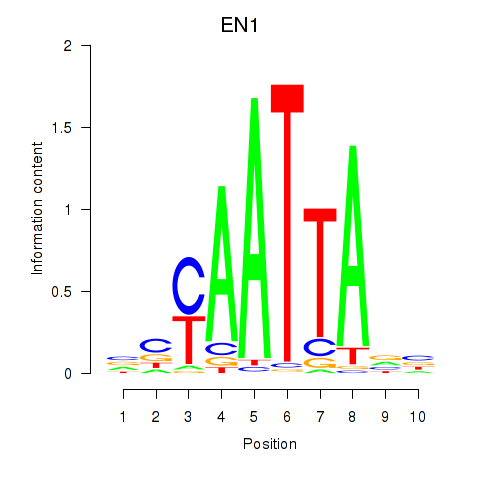
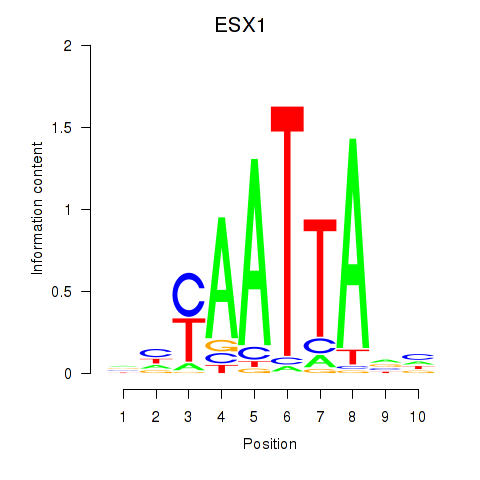
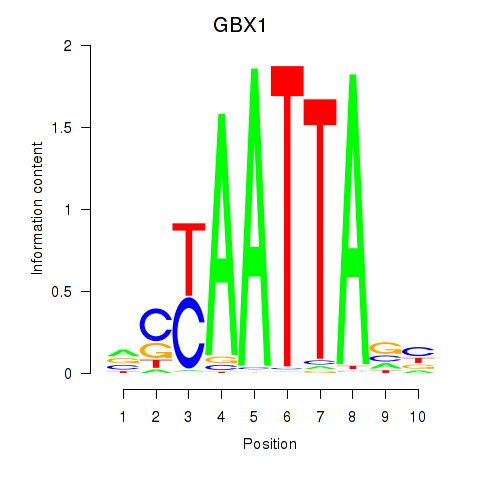
Transcription factors associated with EN1_ESX1_GBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN1
|
ENSG00000163064.6 | EN1 |
|
ESX1
|
ENSG00000123576.5 | ESX1 |
|
GBX1
|
ENSG00000164900.4 | GBX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GBX1 | hg19_v2_chr7_-_150864635_150864785 | -0.20 | 6.3e-01 | Click! |
| ESX1 | hg19_v2_chrX_-_103499602_103499617 | 0.15 | 7.1e-01 | Click! |
| EN1 | hg19_v2_chr2_-_119605253_119605264 | -0.01 | 9.7e-01 | Click! |
Activity profile of EN1_ESX1_GBX1 motif
Sorted Z-values of EN1_ESX1_GBX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EN1_ESX1_GBX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51522955 | 1.13 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr1_-_242612779 | 0.81 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr12_-_28123206 | 0.71 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_28122980 | 0.67 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr5_+_52776449 | 0.54 |
ENST00000396947.3 |
FST |
follistatin |
| chr5_+_52776228 | 0.51 |
ENST00000256759.3 |
FST |
follistatin |
| chr1_+_62439037 | 0.49 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_+_107712173 | 0.47 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr5_+_66300446 | 0.44 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_74486217 | 0.38 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_-_151034734 | 0.37 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr3_+_111718036 | 0.36 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr3_+_111717600 | 0.35 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr3_+_111718173 | 0.34 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr4_+_77356248 | 0.31 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr3_+_111717511 | 0.30 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr12_-_52828147 | 0.30 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr19_-_36001113 | 0.30 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr1_+_153746683 | 0.29 |
ENST00000271857.2 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr1_+_160160346 | 0.28 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr17_-_64225508 | 0.28 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_+_160160283 | 0.27 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr18_+_29027696 | 0.27 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr8_-_41166953 | 0.26 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr4_-_74486347 | 0.25 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_74486109 | 0.24 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_-_82969405 | 0.24 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr4_+_40198527 | 0.23 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr3_+_130569429 | 0.22 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_135394840 | 0.22 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr6_+_130339710 | 0.22 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr17_-_19015945 | 0.21 |
ENST00000573866.2 |
SNORD3D |
small nucleolar RNA, C/D box 3D |
| chr1_+_153747746 | 0.21 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr14_+_22739823 | 0.20 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr10_-_50970322 | 0.20 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr8_+_98900132 | 0.20 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr1_+_160370344 | 0.19 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr4_-_143226979 | 0.19 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr17_-_7167279 | 0.19 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr10_-_50970382 | 0.19 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr8_+_32579341 | 0.19 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr8_+_98881268 | 0.19 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr8_-_86290333 | 0.19 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr14_-_78083112 | 0.19 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr13_-_86373536 | 0.18 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr14_-_38064198 | 0.18 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr20_-_50418947 | 0.18 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr18_+_55888767 | 0.18 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_-_143227088 | 0.18 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr20_-_50418972 | 0.18 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr1_+_28261533 | 0.18 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_+_50300427 | 0.18 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr3_-_160823040 | 0.17 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chrX_+_135730297 | 0.17 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr3_-_160823158 | 0.17 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr18_+_34124507 | 0.17 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr1_+_28261492 | 0.17 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_28261621 | 0.16 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_+_19091325 | 0.16 |
ENST00000584923.1 |
SNORD3A |
small nucleolar RNA, C/D box 3A |
| chr18_-_33709268 | 0.16 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr11_-_117748138 | 0.16 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747434 | 0.16 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747607 | 0.15 |
ENST00000540359.1 ENST00000539526.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chrX_+_134654540 | 0.15 |
ENST00000370752.4 |
DDX26B |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr5_-_141249154 | 0.15 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr12_+_20963647 | 0.15 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_41831485 | 0.15 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr12_-_89746173 | 0.15 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr12_+_20963632 | 0.14 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_46126093 | 0.14 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr10_-_118928543 | 0.14 |
ENST00000419373.2 |
RP11-501J20.2 |
RP11-501J20.2 |
| chr17_-_39341594 | 0.14 |
ENST00000398472.1 |
KRTAP4-1 |
keratin associated protein 4-1 |
| chr9_-_5304432 | 0.14 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr19_-_7968427 | 0.14 |
ENST00000539278.1 |
AC010336.1 |
Uncharacterized protein |
| chr2_+_68961934 | 0.14 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr11_-_124190184 | 0.14 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr2_+_68961905 | 0.14 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr12_+_122688090 | 0.14 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr6_-_111927062 | 0.14 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr17_-_18430160 | 0.13 |
ENST00000392176.3 |
FAM106A |
family with sequence similarity 106, member A |
| chrX_+_107288197 | 0.13 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr8_-_139926236 | 0.13 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr12_+_56473939 | 0.13 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr3_+_121774202 | 0.12 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr11_-_118023490 | 0.12 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr20_-_50419055 | 0.12 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr4_-_116034979 | 0.12 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr2_-_208031943 | 0.12 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr20_+_31805131 | 0.12 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr10_+_94451574 | 0.12 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chrX_+_107288239 | 0.11 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr12_+_44229846 | 0.11 |
ENST00000551577.1 ENST00000266534.3 |
TMEM117 |
transmembrane protein 117 |
| chr3_+_97868170 | 0.11 |
ENST00000437310.1 |
OR5H14 |
olfactory receptor, family 5, subfamily H, member 14 |
| chr19_+_34287751 | 0.11 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr1_-_109935819 | 0.11 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr2_+_159825143 | 0.11 |
ENST00000454300.1 ENST00000263635.6 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr4_-_8873531 | 0.11 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chr5_-_24645078 | 0.11 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr17_-_39324424 | 0.11 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr11_-_128894053 | 0.10 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr21_-_32253874 | 0.10 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chrX_-_130423200 | 0.10 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr1_-_201140673 | 0.10 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr14_-_54423529 | 0.10 |
ENST00000245451.4 ENST00000559087.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr7_-_130080977 | 0.10 |
ENST00000223208.5 |
CEP41 |
centrosomal protein 41kDa |
| chr8_-_40755333 | 0.10 |
ENST00000297737.6 ENST00000315769.7 |
ZMAT4 |
zinc finger, matrin-type 4 |
| chr6_+_29274403 | 0.10 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr12_+_106751436 | 0.10 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr11_+_57435219 | 0.10 |
ENST00000527985.1 ENST00000287169.3 |
ZDHHC5 |
zinc finger, DHHC-type containing 5 |
| chr7_-_100860851 | 0.10 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr12_-_14849470 | 0.10 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr1_-_209979375 | 0.09 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr8_+_38831683 | 0.09 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr17_+_42785976 | 0.09 |
ENST00000393547.2 ENST00000398338.3 |
DBF4B |
DBF4 homolog B (S. cerevisiae) |
| chr19_-_6737576 | 0.09 |
ENST00000601716.1 ENST00000264080.7 |
GPR108 |
G protein-coupled receptor 108 |
| chrX_-_110655306 | 0.09 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr4_-_105416039 | 0.09 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr7_+_73245193 | 0.09 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr3_-_74570291 | 0.09 |
ENST00000263665.6 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
| chr14_+_22670455 | 0.09 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr8_-_20040638 | 0.09 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr17_-_38911580 | 0.09 |
ENST00000312150.4 |
KRT25 |
keratin 25 |
| chr18_+_5748793 | 0.09 |
ENST00000566533.1 ENST00000562452.2 |
RP11-945C19.1 |
RP11-945C19.1 |
| chr21_-_42219065 | 0.09 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chr13_-_36050819 | 0.09 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr1_+_209878182 | 0.09 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr8_-_20040601 | 0.09 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr4_+_71200681 | 0.09 |
ENST00000273936.5 |
CABS1 |
calcium-binding protein, spermatid-specific 1 |
| chrX_-_33229636 | 0.08 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr3_-_98235962 | 0.08 |
ENST00000513873.1 |
CLDND1 |
claudin domain containing 1 |
| chr3_-_55521323 | 0.08 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr5_+_126984710 | 0.08 |
ENST00000379445.3 |
CTXN3 |
cortexin 3 |
| chr17_-_27418537 | 0.08 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_+_117544366 | 0.08 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr16_+_9185450 | 0.08 |
ENST00000327827.7 |
C16orf72 |
chromosome 16 open reading frame 72 |
| chr9_-_107690420 | 0.08 |
ENST00000423487.2 ENST00000374733.1 ENST00000374736.3 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr17_+_47448102 | 0.08 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr11_-_129062093 | 0.08 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr7_+_116660246 | 0.08 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr11_+_34664014 | 0.08 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr4_+_70796784 | 0.08 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr17_+_42786051 | 0.08 |
ENST00000315005.3 |
DBF4B |
DBF4 homolog B (S. cerevisiae) |
| chr19_+_39421556 | 0.08 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr4_-_72649763 | 0.08 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr1_+_118472343 | 0.08 |
ENST00000369441.3 ENST00000349139.5 |
WDR3 |
WD repeat domain 3 |
| chr7_-_33102338 | 0.08 |
ENST00000610140.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr6_-_111927449 | 0.08 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr19_+_13049413 | 0.07 |
ENST00000316448.5 ENST00000588454.1 |
CALR |
calreticulin |
| chr12_-_51422017 | 0.07 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr16_-_29910853 | 0.07 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr10_+_24497704 | 0.07 |
ENST00000376456.4 ENST00000458595.1 |
KIAA1217 |
KIAA1217 |
| chr1_-_235098935 | 0.07 |
ENST00000423175.1 |
RP11-443B7.1 |
RP11-443B7.1 |
| chr10_+_24755416 | 0.07 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr7_-_33102399 | 0.07 |
ENST00000242210.7 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr14_-_82000140 | 0.07 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr3_-_108248169 | 0.07 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr3_-_123339343 | 0.07 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chrX_+_135730373 | 0.07 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr4_+_78432907 | 0.07 |
ENST00000286758.4 |
CXCL13 |
chemokine (C-X-C motif) ligand 13 |
| chr14_+_39734482 | 0.07 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr11_+_34663913 | 0.07 |
ENST00000532302.1 |
EHF |
ets homologous factor |
| chrX_+_49687267 | 0.07 |
ENST00000376091.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr19_+_4007644 | 0.07 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr1_+_244515930 | 0.07 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr1_+_171227069 | 0.07 |
ENST00000354841.4 |
FMO1 |
flavin containing monooxygenase 1 |
| chr3_-_143567262 | 0.07 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr4_-_25865159 | 0.07 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr22_-_32860427 | 0.06 |
ENST00000534972.1 ENST00000397450.1 ENST00000397452.1 |
BPIFC |
BPI fold containing family C |
| chr20_+_43538756 | 0.06 |
ENST00000537323.1 ENST00000217073.2 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chr15_+_58702742 | 0.06 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr5_+_140557371 | 0.06 |
ENST00000239444.2 |
PCDHB8 |
protocadherin beta 8 |
| chr5_-_74162605 | 0.06 |
ENST00000389156.4 ENST00000510496.1 ENST00000380515.3 |
FAM169A |
family with sequence similarity 169, member A |
| chr2_-_99871570 | 0.06 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr4_+_169013666 | 0.06 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr19_-_6737241 | 0.06 |
ENST00000430424.4 ENST00000597298.1 |
GPR108 |
G protein-coupled receptor 108 |
| chr15_+_54901540 | 0.06 |
ENST00000539562.2 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr14_+_68086515 | 0.06 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr19_+_14640372 | 0.06 |
ENST00000215567.5 ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR |
trans-2,3-enoyl-CoA reductase |
| chr6_+_135502501 | 0.06 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_+_81771806 | 0.06 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr20_+_43538692 | 0.06 |
ENST00000217074.4 ENST00000255136.3 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chr6_+_127898312 | 0.06 |
ENST00000329722.7 |
C6orf58 |
chromosome 6 open reading frame 58 |
| chr9_+_136501478 | 0.06 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr2_-_101925055 | 0.06 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr1_+_236958554 | 0.06 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr12_+_16500599 | 0.06 |
ENST00000535309.1 ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chr6_-_31088214 | 0.06 |
ENST00000376288.2 |
CDSN |
corneodesmosin |
| chr6_+_30881982 | 0.06 |
ENST00000321897.5 ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2 |
valyl-tRNA synthetase 2, mitochondrial |
| chr11_-_64684672 | 0.06 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr17_-_39296739 | 0.06 |
ENST00000345847.4 |
KRTAP4-6 |
keratin associated protein 4-6 |
| chr5_+_31193847 | 0.06 |
ENST00000514738.1 ENST00000265071.2 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr12_-_23737534 | 0.06 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chrM_+_12331 | 0.06 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_+_120626919 | 0.06 |
ENST00000273666.6 ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L |
syntaxin binding protein 5-like |
| chr11_+_65554493 | 0.06 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr9_-_5339873 | 0.06 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr19_+_45458503 | 0.06 |
ENST00000337392.5 ENST00000591304.1 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
| chr12_+_16500571 | 0.06 |
ENST00000543076.1 ENST00000396210.3 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chr10_+_24738355 | 0.06 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr14_+_22771851 | 0.06 |
ENST00000390466.1 |
TRAV39 |
T cell receptor alpha variable 39 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 1.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:2001027 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) |
| 0.0 | 0.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.0 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.0 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |