Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for EP300
Z-value: 0.87
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | EP300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | -0.85 | 7.2e-03 | Click! |
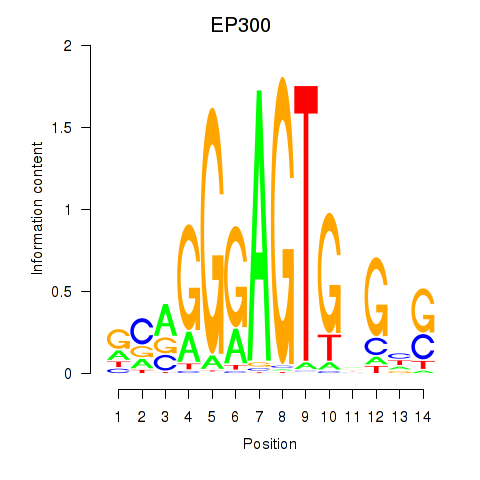
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_107018969 | 0.95 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr10_-_21463116 | 0.77 |
ENST00000417816.2 |
NEBL |
nebulette |
| chr8_+_17434689 | 0.74 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr11_+_15136462 | 0.70 |
ENST00000379556.3 ENST00000424273.1 |
INSC |
inscuteable homolog (Drosophila) |
| chr5_+_119799927 | 0.70 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr11_+_64879317 | 0.69 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr12_-_54867352 | 0.62 |
ENST00000305879.5 |
GTSF1 |
gametocyte specific factor 1 |
| chr13_-_44361025 | 0.57 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chr19_+_12862604 | 0.57 |
ENST00000553030.1 |
BEST2 |
bestrophin 2 |
| chrX_-_107019181 | 0.56 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr14_+_105939276 | 0.54 |
ENST00000483017.3 |
CRIP2 |
cysteine-rich protein 2 |
| chr19_+_12862486 | 0.53 |
ENST00000549706.1 |
BEST2 |
bestrophin 2 |
| chr10_-_81205373 | 0.51 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr14_+_52327109 | 0.48 |
ENST00000335281.4 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_-_49812997 | 0.47 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_211432700 | 0.40 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr5_+_139028510 | 0.39 |
ENST00000502336.1 ENST00000520967.1 ENST00000511048.1 |
CXXC5 |
CXXC finger protein 5 |
| chrX_-_140271249 | 0.38 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr22_-_29075853 | 0.37 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr19_-_50311896 | 0.37 |
ENST00000529634.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr20_-_30311703 | 0.35 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr15_-_82338460 | 0.34 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr2_+_24346324 | 0.34 |
ENST00000407625.1 ENST00000420135.2 |
FAM228B |
family with sequence similarity 228, member B |
| chr7_-_150974494 | 0.32 |
ENST00000392811.2 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr3_+_151986709 | 0.32 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr17_-_19266045 | 0.31 |
ENST00000395616.3 |
B9D1 |
B9 protein domain 1 |
| chr12_-_105630016 | 0.30 |
ENST00000258530.3 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr1_-_161102367 | 0.29 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr10_-_49813090 | 0.29 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr10_+_104178946 | 0.28 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr2_-_160919112 | 0.28 |
ENST00000283243.7 ENST00000392771.1 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
| chr6_+_2988847 | 0.28 |
ENST00000380472.3 ENST00000605901.1 ENST00000454015.1 |
NQO2 LINC01011 |
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr6_+_3000057 | 0.28 |
ENST00000397717.2 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr6_+_3000195 | 0.28 |
ENST00000338130.2 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr2_-_203735976 | 0.27 |
ENST00000435143.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr19_-_45927097 | 0.27 |
ENST00000340192.7 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr5_-_39425068 | 0.27 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_7080227 | 0.27 |
ENST00000574330.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr2_+_74154032 | 0.27 |
ENST00000356837.6 |
DGUOK |
deoxyguanosine kinase |
| chr17_+_47075023 | 0.27 |
ENST00000431824.2 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr1_-_2126192 | 0.26 |
ENST00000378546.4 |
C1orf86 |
chromosome 1 open reading frame 86 |
| chr17_-_7145475 | 0.26 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr1_+_154300217 | 0.26 |
ENST00000368489.3 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr22_-_41985865 | 0.26 |
ENST00000216259.7 |
PMM1 |
phosphomannomutase 1 |
| chr1_+_3541543 | 0.25 |
ENST00000378344.2 ENST00000344579.5 |
TPRG1L |
tumor protein p63 regulated 1-like |
| chrX_+_134166333 | 0.25 |
ENST00000257013.7 |
FAM127A |
family with sequence similarity 127, member A |
| chr12_-_31744031 | 0.25 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr20_-_33460621 | 0.25 |
ENST00000427420.1 ENST00000336431.5 |
GGT7 |
gamma-glutamyltransferase 7 |
| chr4_+_62066941 | 0.25 |
ENST00000512091.2 |
LPHN3 |
latrophilin 3 |
| chr12_-_105629852 | 0.25 |
ENST00000551662.1 ENST00000553097.1 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr11_+_75428857 | 0.25 |
ENST00000198801.5 |
MOGAT2 |
monoacylglycerol O-acyltransferase 2 |
| chr6_+_3000218 | 0.25 |
ENST00000380441.1 ENST00000380455.4 ENST00000380454.4 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr10_+_45495898 | 0.25 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr2_+_74153953 | 0.25 |
ENST00000264093.4 ENST00000348222.1 |
DGUOK |
deoxyguanosine kinase |
| chr6_-_34360413 | 0.25 |
ENST00000607016.1 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr2_-_27294500 | 0.24 |
ENST00000447619.1 ENST00000429985.1 ENST00000456793.1 |
OST4 |
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr11_-_64527425 | 0.24 |
ENST00000377432.3 |
PYGM |
phosphorylase, glycogen, muscle |
| chr5_-_39425222 | 0.24 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_+_81892347 | 0.23 |
ENST00000372267.2 |
PLAC9 |
placenta-specific 9 |
| chr6_-_41703296 | 0.23 |
ENST00000373033.1 |
TFEB |
transcription factor EB |
| chr19_+_51226573 | 0.23 |
ENST00000250340.4 |
CLEC11A |
C-type lectin domain family 11, member A |
| chrX_+_55478538 | 0.23 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr8_+_144816303 | 0.23 |
ENST00000533004.1 |
FAM83H-AS1 |
FAM83H antisense RNA 1 (head to head) |
| chr10_+_76585303 | 0.23 |
ENST00000372725.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr5_-_39425290 | 0.22 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_54947229 | 0.22 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr8_-_91657909 | 0.21 |
ENST00000418210.2 |
TMEM64 |
transmembrane protein 64 |
| chr17_+_37783170 | 0.21 |
ENST00000254079.4 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_109656439 | 0.21 |
ENST00000369949.4 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr3_-_171528227 | 0.21 |
ENST00000356327.5 ENST00000342215.6 ENST00000340989.4 ENST00000351298.4 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
| chr1_-_10003372 | 0.21 |
ENST00000377223.1 ENST00000541052.1 ENST00000377213.1 |
LZIC |
leucine zipper and CTNNBIP1 domain containing |
| chr17_-_79995553 | 0.21 |
ENST00000581584.1 ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR |
dicarbonyl/L-xylulose reductase |
| chr17_+_66511540 | 0.20 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr20_-_33735070 | 0.20 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr17_+_39969183 | 0.20 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr12_+_4699244 | 0.20 |
ENST00000540757.2 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr11_-_114271139 | 0.20 |
ENST00000325636.4 |
C11orf71 |
chromosome 11 open reading frame 71 |
| chr17_-_19265855 | 0.20 |
ENST00000440841.1 ENST00000395615.1 ENST00000461069.2 |
B9D1 |
B9 protein domain 1 |
| chr18_+_5238549 | 0.20 |
ENST00000580684.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr17_-_19265982 | 0.20 |
ENST00000268841.6 ENST00000261499.4 ENST00000575478.1 |
B9D1 |
B9 protein domain 1 |
| chr2_+_220436917 | 0.20 |
ENST00000243786.2 |
INHA |
inhibin, alpha |
| chr22_+_42475692 | 0.20 |
ENST00000331479.3 |
SMDT1 |
single-pass membrane protein with aspartate-rich tail 1 |
| chr19_-_49314169 | 0.19 |
ENST00000597011.1 ENST00000601681.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr2_+_74685413 | 0.19 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr2_+_74685527 | 0.19 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chr12_-_31743901 | 0.19 |
ENST00000354285.4 |
DENND5B |
DENN/MADD domain containing 5B |
| chr9_+_19049372 | 0.19 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr9_-_139760717 | 0.19 |
ENST00000371648.4 |
EDF1 |
endothelial differentiation-related factor 1 |
| chr12_+_51318513 | 0.19 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr20_-_44485835 | 0.18 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr19_-_49314269 | 0.18 |
ENST00000545387.2 ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr8_+_27168988 | 0.18 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr7_+_45197383 | 0.18 |
ENST00000242249.4 ENST00000496212.1 ENST00000481345.1 |
RAMP3 |
receptor (G protein-coupled) activity modifying protein 3 |
| chrX_+_153770421 | 0.18 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr18_+_48494361 | 0.18 |
ENST00000588577.1 ENST00000269466.3 ENST00000591429.1 ENST00000452201.2 |
ELAC1 SMAD4 |
elaC ribonuclease Z 1 SMAD family member 4 |
| chr1_+_202431859 | 0.18 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_77582905 | 0.18 |
ENST00000557408.1 |
TMEM63C |
transmembrane protein 63C |
| chr1_+_211433275 | 0.18 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr14_-_65346555 | 0.18 |
ENST00000542895.1 ENST00000556626.1 |
SPTB |
spectrin, beta, erythrocytic |
| chr2_+_30369807 | 0.18 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr1_+_6845578 | 0.18 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr16_-_27279900 | 0.18 |
ENST00000564342.1 ENST00000567710.1 ENST00000563273.1 ENST00000361439.4 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
| chr19_-_56109119 | 0.17 |
ENST00000587678.1 |
FIZ1 |
FLT3-interacting zinc finger 1 |
| chr2_-_24346218 | 0.17 |
ENST00000436622.1 ENST00000313213.4 |
PFN4 |
profilin family, member 4 |
| chr8_-_91658303 | 0.17 |
ENST00000458549.2 |
TMEM64 |
transmembrane protein 64 |
| chr21_-_38639601 | 0.17 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr18_-_5238525 | 0.17 |
ENST00000581170.1 ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5 LINC00526 |
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr4_-_142053952 | 0.17 |
ENST00000515673.2 |
RNF150 |
ring finger protein 150 |
| chr17_-_74099772 | 0.17 |
ENST00000411744.2 ENST00000332065.5 |
EXOC7 |
exocyst complex component 7 |
| chr1_+_36771946 | 0.17 |
ENST00000373139.2 ENST00000453908.2 ENST00000426732.2 |
SH3D21 |
SH3 domain containing 21 |
| chr2_-_233877912 | 0.17 |
ENST00000264051.3 |
NGEF |
neuronal guanine nucleotide exchange factor |
| chr10_-_32636106 | 0.17 |
ENST00000263062.8 ENST00000319778.6 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
| chr11_+_105481612 | 0.17 |
ENST00000531011.1 ENST00000525187.1 ENST00000530497.1 |
GRIA4 |
glutamate receptor, ionotropic, AMPA 4 |
| chr1_+_6845497 | 0.17 |
ENST00000473578.1 ENST00000557126.1 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr14_-_89021077 | 0.16 |
ENST00000556564.1 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
| chr16_+_47495201 | 0.16 |
ENST00000566044.1 ENST00000455779.1 |
PHKB |
phosphorylase kinase, beta |
| chr2_+_30369859 | 0.16 |
ENST00000402003.3 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr8_-_27630102 | 0.16 |
ENST00000356537.4 ENST00000522915.1 ENST00000539095.1 |
CCDC25 |
coiled-coil domain containing 25 |
| chr16_+_28889703 | 0.16 |
ENST00000357084.3 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr15_+_76196234 | 0.16 |
ENST00000540507.1 ENST00000565036.1 ENST00000569054.1 |
FBXO22 |
F-box protein 22 |
| chr5_-_65017921 | 0.16 |
ENST00000381007.4 |
SGTB |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chrX_-_71792477 | 0.16 |
ENST00000421523.1 ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8 |
histone deacetylase 8 |
| chr19_+_51226648 | 0.16 |
ENST00000599973.1 |
CLEC11A |
C-type lectin domain family 11, member A |
| chr16_+_47495225 | 0.16 |
ENST00000299167.8 ENST00000323584.5 ENST00000563376.1 |
PHKB |
phosphorylase kinase, beta |
| chr7_-_5569588 | 0.16 |
ENST00000417101.1 |
ACTB |
actin, beta |
| chr19_+_11909329 | 0.16 |
ENST00000323169.5 ENST00000450087.1 |
ZNF491 |
zinc finger protein 491 |
| chr10_-_121302195 | 0.15 |
ENST00000369103.2 |
RGS10 |
regulator of G-protein signaling 10 |
| chr12_+_54378923 | 0.15 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr5_+_43603229 | 0.15 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr22_-_43042955 | 0.15 |
ENST00000402438.1 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr9_-_139760737 | 0.15 |
ENST00000371649.1 ENST00000224073.1 |
EDF1 |
endothelial differentiation-related factor 1 |
| chr22_-_31688381 | 0.15 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr19_+_49617581 | 0.15 |
ENST00000391864.3 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr19_-_18649181 | 0.15 |
ENST00000596015.1 |
FKBP8 |
FK506 binding protein 8, 38kDa |
| chr4_-_114682224 | 0.15 |
ENST00000342666.5 ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_161102421 | 0.15 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr2_-_204400013 | 0.15 |
ENST00000374489.2 ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_+_101917407 | 0.15 |
ENST00000487284.1 |
CUX1 |
cut-like homeobox 1 |
| chr21_+_45285050 | 0.15 |
ENST00000291572.8 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr17_-_74099795 | 0.14 |
ENST00000406660.3 ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7 |
exocyst complex component 7 |
| chrX_+_135229600 | 0.14 |
ENST00000370690.3 |
FHL1 |
four and a half LIM domains 1 |
| chr14_-_61190754 | 0.14 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr4_-_114682364 | 0.14 |
ENST00000511664.1 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr3_+_105086056 | 0.14 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chrX_+_135229559 | 0.14 |
ENST00000394155.2 |
FHL1 |
four and a half LIM domains 1 |
| chr10_-_104192405 | 0.14 |
ENST00000369937.4 |
CUEDC2 |
CUE domain containing 2 |
| chrX_-_70474910 | 0.14 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr1_-_161102213 | 0.14 |
ENST00000458050.2 |
DEDD |
death effector domain containing |
| chr14_-_77923897 | 0.14 |
ENST00000343765.2 ENST00000327028.4 ENST00000556412.1 ENST00000557466.1 ENST00000448935.2 ENST00000553888.1 ENST00000557658.1 |
VIPAS39 |
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr9_-_123638633 | 0.14 |
ENST00000456291.1 |
PHF19 |
PHD finger protein 19 |
| chrX_-_100872911 | 0.14 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr19_+_19626531 | 0.14 |
ENST00000507754.4 |
NDUFA13 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr16_+_69458428 | 0.14 |
ENST00000512062.1 ENST00000307892.8 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
| chr22_+_20105012 | 0.14 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr18_-_21977748 | 0.14 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr1_-_19229014 | 0.14 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr2_-_110371664 | 0.13 |
ENST00000545389.1 ENST00000423520.1 |
SEPT10 |
septin 10 |
| chr11_+_1968508 | 0.13 |
ENST00000397298.3 ENST00000381519.1 ENST00000397297.3 ENST00000381514.3 ENST00000397294.3 |
MRPL23 |
mitochondrial ribosomal protein L23 |
| chr22_+_42470244 | 0.13 |
ENST00000321753.3 |
FAM109B |
family with sequence similarity 109, member B |
| chr10_-_102746953 | 0.13 |
ENST00000523148.1 |
MRPL43 |
mitochondrial ribosomal protein L43 |
| chr16_-_66968265 | 0.13 |
ENST00000567511.1 ENST00000422424.2 |
FAM96B |
family with sequence similarity 96, member B |
| chr2_-_99224915 | 0.13 |
ENST00000328709.3 ENST00000409997.1 |
COA5 |
cytochrome c oxidase assembly factor 5 |
| chr17_-_71308119 | 0.13 |
ENST00000439510.2 ENST00000581014.1 ENST00000579611.1 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
| chr20_+_32399093 | 0.13 |
ENST00000217402.2 |
CHMP4B |
charged multivesicular body protein 4B |
| chr16_+_2083265 | 0.13 |
ENST00000565855.1 ENST00000566198.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr1_+_236558694 | 0.13 |
ENST00000359362.5 |
EDARADD |
EDAR-associated death domain |
| chr2_-_153032484 | 0.13 |
ENST00000263904.4 |
STAM2 |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr19_+_1000418 | 0.13 |
ENST00000234389.3 |
GRIN3B |
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr19_+_16296191 | 0.13 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr4_-_114682597 | 0.13 |
ENST00000394524.3 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr22_+_20105259 | 0.13 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr3_-_71632894 | 0.12 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chr22_-_43042968 | 0.12 |
ENST00000407623.3 ENST00000396303.3 ENST00000438270.1 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr19_+_49617609 | 0.12 |
ENST00000221459.2 ENST00000486217.2 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr15_+_38544476 | 0.12 |
ENST00000299084.4 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
| chr11_-_100999775 | 0.12 |
ENST00000263463.5 |
PGR |
progesterone receptor |
| chr18_-_34408902 | 0.12 |
ENST00000593035.1 ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr10_-_64028466 | 0.12 |
ENST00000395265.1 ENST00000373789.3 ENST00000395260.3 |
RTKN2 |
rhotekin 2 |
| chr1_+_222886694 | 0.12 |
ENST00000426638.1 ENST00000537020.1 ENST00000539697.1 |
BROX |
BRO1 domain and CAAX motif containing |
| chr11_-_1771797 | 0.12 |
ENST00000340134.4 |
IFITM10 |
interferon induced transmembrane protein 10 |
| chr9_+_131218698 | 0.12 |
ENST00000434106.3 ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr5_+_162864575 | 0.12 |
ENST00000512163.1 ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1 |
cyclin G1 |
| chr2_-_25016251 | 0.12 |
ENST00000328379.5 |
PTRHD1 |
peptidyl-tRNA hydrolase domain containing 1 |
| chr14_-_57735528 | 0.12 |
ENST00000340918.7 ENST00000413566.2 |
EXOC5 |
exocyst complex component 5 |
| chr10_-_105212141 | 0.12 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr16_-_66968055 | 0.12 |
ENST00000568572.1 |
FAM96B |
family with sequence similarity 96, member B |
| chr5_+_36152179 | 0.12 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_-_2126174 | 0.12 |
ENST00000400919.3 ENST00000420515.1 ENST00000378543.2 ENST00000400918.3 |
C1orf86 |
chromosome 1 open reading frame 86 |
| chr10_-_98480243 | 0.12 |
ENST00000339364.5 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
| chr21_+_17102311 | 0.12 |
ENST00000285679.6 ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25 |
ubiquitin specific peptidase 25 |
| chr1_-_43855479 | 0.12 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr6_+_122793058 | 0.11 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr2_+_233415488 | 0.11 |
ENST00000454501.1 |
EIF4E2 |
eukaryotic translation initiation factor 4E family member 2 |
| chr2_+_87808725 | 0.11 |
ENST00000413202.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr19_-_36231437 | 0.11 |
ENST00000591748.1 |
IGFLR1 |
IGF-like family receptor 1 |
| chr12_+_27677085 | 0.11 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr18_-_34408802 | 0.11 |
ENST00000590842.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr11_-_18610246 | 0.11 |
ENST00000379387.4 ENST00000541984.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr6_-_149969829 | 0.11 |
ENST00000367411.2 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr7_-_2354099 | 0.11 |
ENST00000222990.3 |
SNX8 |
sorting nexin 8 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 1.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.5 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.4 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 1.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.7 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.4 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0061366 | olfactory nerve development(GO:0021553) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.2 | 0.7 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.7 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.4 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |