Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
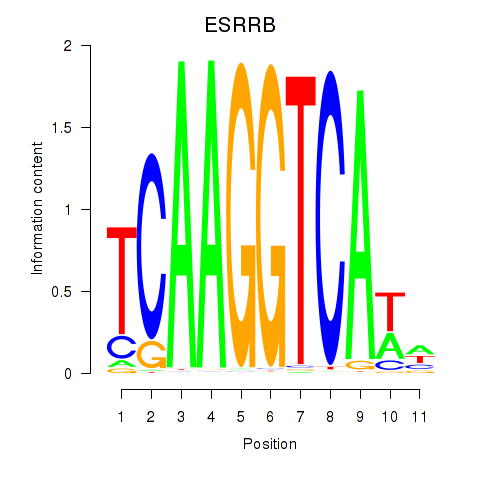
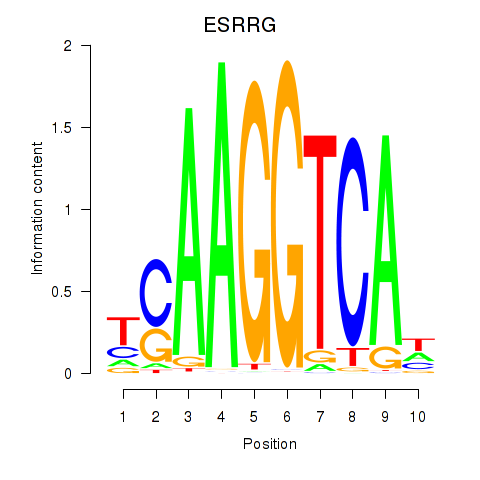
Results for ESRRB_ESRRG
Z-value: 1.74
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | ESRRB |
|
ESRRG
|
ENSG00000196482.12 | ESRRG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRB | hg19_v2_chr14_+_76776957_76777061 | 0.80 | 1.6e-02 | Click! |
| ESRRG | hg19_v2_chr1_-_217262969_217262995, hg19_v2_chr1_-_217311090_217311097, hg19_v2_chr1_-_217262933_217262968, hg19_v2_chr1_-_216896780_216896800, hg19_v2_chr1_-_216978709_216978728 | 0.77 | 2.6e-02 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
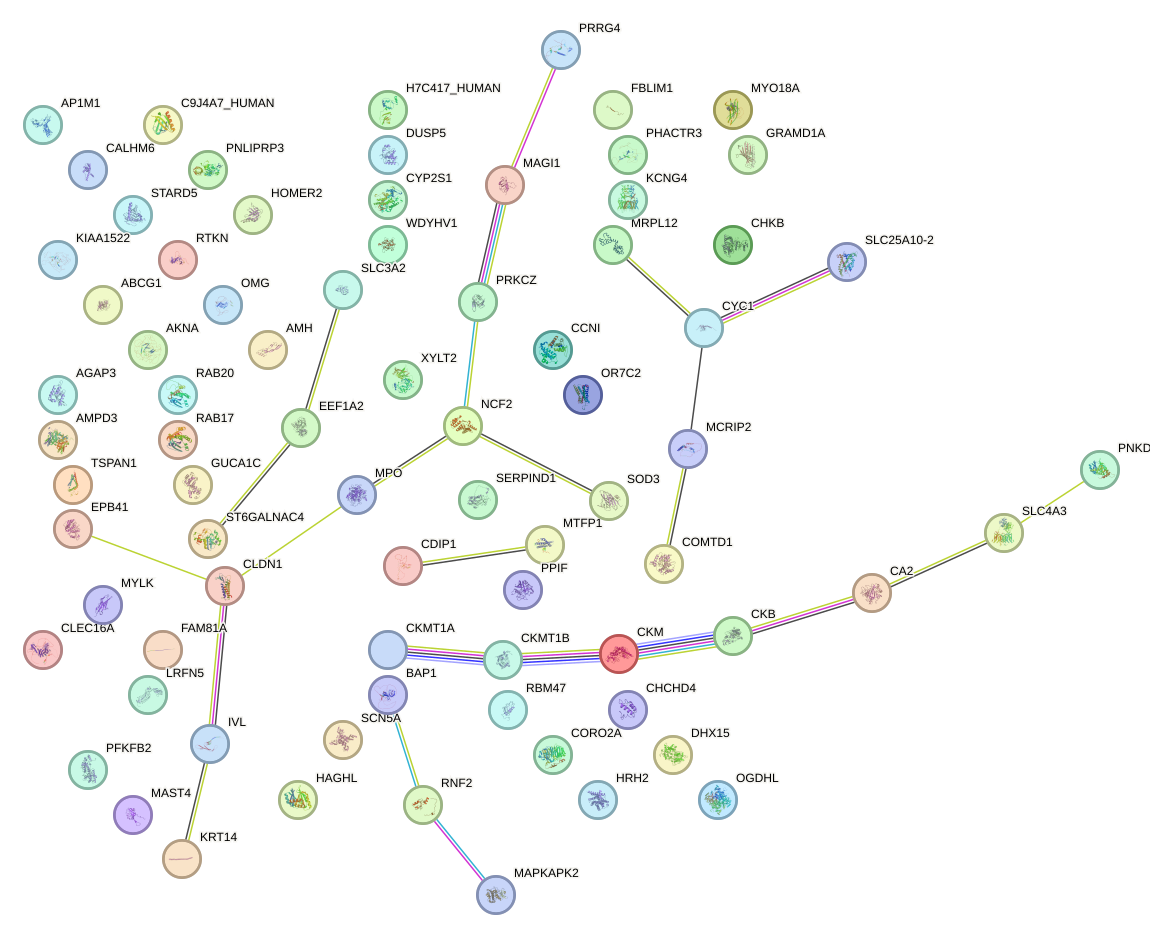
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_58179582 | 2.97 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr2_+_220491973 | 2.81 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr15_+_43886057 | 2.55 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43885252 | 2.42 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985725 | 2.35 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43985084 | 2.34 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr8_+_86376081 | 2.32 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr12_-_53320245 | 2.30 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr2_-_235405168 | 2.19 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr2_+_220492116 | 2.15 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr1_+_46640750 | 1.82 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr20_+_58152524 | 1.77 |
ENST00000359926.3 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr2_+_220492287 | 1.44 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr5_-_60140009 | 1.40 |
ENST00000505959.1 |
ELOVL7 |
ELOVL fatty acid elongase 7 |
| chr2_+_220492373 | 1.37 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr10_+_118187379 | 1.36 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr15_-_83621435 | 1.33 |
ENST00000450735.2 ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2 |
homer homolog 2 (Drosophila) |
| chr9_-_117150243 | 1.21 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr3_+_113666748 | 1.19 |
ENST00000330212.3 ENST00000498275.1 |
ZDHHC23 |
zinc finger, DHHC-type containing 23 |
| chr10_-_76995769 | 1.12 |
ENST00000372538.3 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr10_-_76995675 | 1.11 |
ENST00000469299.1 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr17_-_27503770 | 1.03 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr11_+_10472223 | 1.02 |
ENST00000396554.3 ENST00000524866.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr1_+_2036149 | 1.01 |
ENST00000482686.1 ENST00000400920.1 ENST00000486681.1 |
PRKCZ |
protein kinase C, zeta |
| chr10_+_81107271 | 0.95 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr18_-_24128496 | 0.88 |
ENST00000417602.1 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chr4_-_40517984 | 0.88 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr15_+_59730348 | 0.86 |
ENST00000288228.5 ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A |
family with sequence similarity 81, member A |
| chr1_+_33231268 | 0.84 |
ENST00000373480.1 |
KIAA1522 |
KIAA1522 |
| chr20_+_1875942 | 0.76 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr10_+_81107216 | 0.75 |
ENST00000394579.3 ENST00000225174.3 |
PPIF |
peptidylprolyl isomerase F |
| chr16_-_4588469 | 0.71 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr3_-_190040223 | 0.68 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr14_-_21491477 | 0.68 |
ENST00000298684.5 ENST00000557169.1 ENST00000553563.1 |
NDRG2 |
NDRG family member 2 |
| chr15_-_51535208 | 0.67 |
ENST00000405913.3 ENST00000559878.1 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr11_+_32851487 | 0.64 |
ENST00000257836.3 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr10_-_50970322 | 0.62 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr10_-_50970382 | 0.61 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr5_+_80529104 | 0.61 |
ENST00000254035.4 ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr17_-_29624343 | 0.60 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr16_+_691792 | 0.58 |
ENST00000307650.4 |
FAM195A |
family with sequence similarity 195, member A |
| chr21_+_43639211 | 0.56 |
ENST00000450121.1 ENST00000398449.3 ENST00000361802.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr1_+_152881014 | 0.56 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr19_-_10697895 | 0.56 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr6_-_33548006 | 0.55 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr8_-_131028660 | 0.55 |
ENST00000401979.2 ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr17_-_39928106 | 0.54 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr16_+_777118 | 0.54 |
ENST00000562141.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr10_+_112257596 | 0.54 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr1_-_183560011 | 0.54 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr16_-_84273304 | 0.54 |
ENST00000308251.4 ENST00000568181.1 |
KCNG4 |
potassium voltage-gated channel, subfamily G, member 4 |
| chr1_+_207226574 | 0.53 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr8_-_131028641 | 0.53 |
ENST00000523509.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr6_-_33547975 | 0.53 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr1_-_183559693 | 0.53 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr2_-_238499337 | 0.52 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr2_-_238499303 | 0.52 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_-_28122980 | 0.51 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr16_+_776936 | 0.50 |
ENST00000549114.1 ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr1_+_16083154 | 0.50 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr19_+_35485682 | 0.49 |
ENST00000599564.1 |
GRAMD1A |
GRAM domain containing 1A |
| chr2_-_238499725 | 0.48 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr4_+_24797085 | 0.48 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr20_-_62130474 | 0.48 |
ENST00000217182.3 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr13_-_111214015 | 0.48 |
ENST00000267328.3 |
RAB20 |
RAB20, member RAS oncogene family |
| chr2_-_238499131 | 0.48 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr8_+_99956759 | 0.47 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr22_+_30821732 | 0.47 |
ENST00000355143.4 |
MTFP1 |
mitochondrial fission process 1 |
| chr3_-_52443799 | 0.46 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr19_+_15052301 | 0.45 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr3_-_123339343 | 0.45 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr15_-_81616446 | 0.44 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr17_+_79670386 | 0.43 |
ENST00000333676.3 ENST00000571730.1 ENST00000541223.1 |
MRPL12 SLC25A10 SLC25A10 |
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr2_-_74667612 | 0.42 |
ENST00000305557.5 ENST00000233330.6 |
RTKN |
rhotekin |
| chr3_-_14166316 | 0.42 |
ENST00000396914.3 ENST00000295767.5 |
CHCHD4 |
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr8_-_131028869 | 0.41 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr16_+_11038403 | 0.40 |
ENST00000409552.3 |
CLEC16A |
C-type lectin domain family 16, member A |
| chr4_-_143227088 | 0.40 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr2_+_219135115 | 0.39 |
ENST00000248451.3 ENST00000273077.4 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
| chr19_+_2249308 | 0.39 |
ENST00000592877.1 ENST00000221496.4 |
AMH |
anti-Mullerian hormone |
| chr5_+_145317356 | 0.39 |
ENST00000511217.1 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr3_-_123339418 | 0.39 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr22_+_21128167 | 0.39 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr11_+_62623621 | 0.38 |
ENST00000535296.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_62623544 | 0.38 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr9_-_130679257 | 0.38 |
ENST00000361444.3 ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr17_-_2614927 | 0.38 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chrX_-_46187069 | 0.37 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr1_+_29213584 | 0.37 |
ENST00000343067.4 ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr14_-_103987679 | 0.37 |
ENST00000553610.1 |
CKB |
creatine kinase, brain |
| chr11_+_62623512 | 0.36 |
ENST00000377892.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr15_-_67813924 | 0.36 |
ENST00000559298.1 |
IQCH-AS1 |
IQCH antisense RNA 1 |
| chr6_+_116782527 | 0.35 |
ENST00000368606.3 ENST00000368605.1 |
FAM26F |
family with sequence similarity 26, member F |
| chr5_+_175085033 | 0.35 |
ENST00000377291.2 |
HRH2 |
histamine receptor H2 |
| chr9_-_100954910 | 0.35 |
ENST00000375077.4 |
CORO2A |
coronin, actin binding protein, 2A |
| chr11_-_66139199 | 0.34 |
ENST00000357440.2 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr7_+_150782945 | 0.34 |
ENST00000463381.1 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr22_+_46546494 | 0.34 |
ENST00000396000.2 ENST00000262735.5 ENST00000420804.1 |
PPARA |
peroxisome proliferator-activated receptor alpha |
| chr8_+_145149930 | 0.34 |
ENST00000318911.4 |
CYC1 |
cytochrome c-1 |
| chr3_-_38691119 | 0.34 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr19_+_41699135 | 0.34 |
ENST00000542619.1 ENST00000600561.1 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr3_-_108672742 | 0.34 |
ENST00000261047.3 |
GUCA1C |
guanylate cyclase activator 1C |
| chr14_-_21490958 | 0.34 |
ENST00000554104.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_21492113 | 0.33 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr17_-_39743139 | 0.33 |
ENST00000167586.6 |
KRT14 |
keratin 14 |
| chr14_-_21492251 | 0.33 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr2_-_237412331 | 0.33 |
ENST00000418802.1 |
IQCA1 |
IQ motif containing with AAA domain 1 |
| chr17_-_56350797 | 0.32 |
ENST00000577220.1 |
MPO |
myeloperoxidase |
| chr6_-_107436473 | 0.32 |
ENST00000369042.1 |
BEND3 |
BEN domain containing 3 |
| chr1_+_206858328 | 0.32 |
ENST00000367103.3 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr5_+_66254698 | 0.31 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_48423453 | 0.31 |
ENST00000017003.2 ENST00000509778.1 ENST00000507602.1 |
XYLT2 |
xylosyltransferase II |
| chr7_+_35756092 | 0.31 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr7_+_35756186 | 0.30 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr14_+_42076765 | 0.30 |
ENST00000298119.4 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
| chr3_-_108672609 | 0.30 |
ENST00000393963.3 ENST00000471108.1 |
GUCA1C |
guanylate cyclase activator 1C |
| chr8_+_124429006 | 0.30 |
ENST00000522194.1 ENST00000523356.1 |
WDYHV1 |
WDYHV motif containing 1 |
| chr15_+_65204075 | 0.30 |
ENST00000380230.3 ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A |
ankyrin repeat and death domain containing 1A |
| chr17_-_26876350 | 0.30 |
ENST00000470125.1 |
UNC119 |
unc-119 homolog (C. elegans) |
| chr8_-_110346614 | 0.30 |
ENST00000239690.4 |
NUDCD1 |
NudC domain containing 1 |
| chr6_-_43027105 | 0.30 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr14_+_22985251 | 0.30 |
ENST00000390510.1 |
TRAJ27 |
T cell receptor alpha joining 27 |
| chr21_-_46330545 | 0.29 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_198365095 | 0.29 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr19_-_2328572 | 0.29 |
ENST00000252622.10 |
LSM7 |
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr18_+_55816546 | 0.29 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_-_156840776 | 0.29 |
ENST00000471357.1 |
LINC00880 |
long intergenic non-protein coding RNA 880 |
| chr15_+_67814008 | 0.29 |
ENST00000557807.1 |
C15orf61 |
chromosome 15 open reading frame 61 |
| chr9_-_118687405 | 0.28 |
ENST00000374014.3 |
LINC00474 |
long intergenic non-protein coding RNA 474 |
| chr1_+_153388993 | 0.28 |
ENST00000368729.4 |
S100A7A |
S100 calcium binding protein A7A |
| chr12_+_10366016 | 0.28 |
ENST00000546017.1 ENST00000535576.1 ENST00000539170.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr7_+_48128854 | 0.28 |
ENST00000436673.1 ENST00000429491.2 |
UPP1 |
uridine phosphorylase 1 |
| chr6_-_43197189 | 0.28 |
ENST00000509253.1 ENST00000393987.2 ENST00000230431.6 |
DNPH1 |
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr20_-_3748363 | 0.28 |
ENST00000217195.8 |
C20orf27 |
chromosome 20 open reading frame 27 |
| chr17_-_6616678 | 0.28 |
ENST00000381074.4 ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr20_-_3748416 | 0.28 |
ENST00000399672.1 |
C20orf27 |
chromosome 20 open reading frame 27 |
| chr15_+_67813406 | 0.28 |
ENST00000342683.4 |
C15orf61 |
chromosome 15 open reading frame 61 |
| chr7_+_48128816 | 0.28 |
ENST00000395564.4 |
UPP1 |
uridine phosphorylase 1 |
| chr9_-_138853156 | 0.28 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr8_-_100905363 | 0.27 |
ENST00000524245.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr1_+_29213678 | 0.27 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr2_-_209028300 | 0.27 |
ENST00000304502.4 |
CRYGA |
crystallin, gamma A |
| chr2_+_238875597 | 0.27 |
ENST00000272930.4 ENST00000448502.1 ENST00000416292.1 ENST00000409633.1 ENST00000414443.1 ENST00000409953.1 ENST00000409332.1 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
| chr17_-_73505961 | 0.26 |
ENST00000433559.2 |
CASKIN2 |
CASK interacting protein 2 |
| chr11_-_72463421 | 0.26 |
ENST00000393609.3 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr22_-_29711645 | 0.26 |
ENST00000401450.3 |
RASL10A |
RAS-like, family 10, member A |
| chr13_+_97928395 | 0.26 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr2_+_192141611 | 0.26 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr4_-_170947522 | 0.26 |
ENST00000361618.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr8_-_100905925 | 0.26 |
ENST00000518171.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr5_+_34757309 | 0.26 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr7_+_48128194 | 0.26 |
ENST00000416681.1 ENST00000331803.4 ENST00000432131.1 |
UPP1 |
uridine phosphorylase 1 |
| chr7_+_48128316 | 0.26 |
ENST00000341253.4 |
UPP1 |
uridine phosphorylase 1 |
| chr17_-_7991021 | 0.26 |
ENST00000319144.4 |
ALOX12B |
arachidonate 12-lipoxygenase, 12R type |
| chr15_+_78441663 | 0.25 |
ENST00000299518.2 ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_-_25256368 | 0.25 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr16_-_3767551 | 0.25 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr22_-_29711704 | 0.25 |
ENST00000216101.6 |
RASL10A |
RAS-like, family 10, member A |
| chr19_-_11689752 | 0.25 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr2_+_120189422 | 0.25 |
ENST00000306406.4 |
TMEM37 |
transmembrane protein 37 |
| chr8_-_100905850 | 0.25 |
ENST00000520271.1 ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr16_+_30937213 | 0.25 |
ENST00000427128.1 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
| chr12_+_120875910 | 0.24 |
ENST00000551806.1 |
AL021546.6 |
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr7_-_151433342 | 0.24 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_151433393 | 0.24 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr16_-_3767506 | 0.23 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr10_-_120925054 | 0.23 |
ENST00000419372.1 ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4 |
sideroflexin 4 |
| chr19_-_55574538 | 0.23 |
ENST00000415061.3 |
RDH13 |
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr19_+_41698927 | 0.23 |
ENST00000310054.4 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr7_+_56032270 | 0.23 |
ENST00000322090.3 ENST00000446778.1 |
GBAS |
glioblastoma amplified sequence |
| chr19_+_1407733 | 0.23 |
ENST00000592453.1 |
DAZAP1 |
DAZ associated protein 1 |
| chr11_-_132813566 | 0.23 |
ENST00000331898.7 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
| chr8_+_99956662 | 0.23 |
ENST00000523368.1 ENST00000297565.4 ENST00000435298.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr4_-_122872909 | 0.22 |
ENST00000379645.3 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr1_-_153433120 | 0.22 |
ENST00000368723.3 |
S100A7 |
S100 calcium binding protein A7 |
| chr11_+_6866883 | 0.22 |
ENST00000299454.4 ENST00000379831.2 |
OR10A5 |
olfactory receptor, family 10, subfamily A, member 5 |
| chrX_-_154033661 | 0.22 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr20_-_1373606 | 0.22 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr19_+_35629702 | 0.22 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr21_+_42539701 | 0.22 |
ENST00000330333.6 ENST00000328735.6 ENST00000347667.5 |
BACE2 |
beta-site APP-cleaving enzyme 2 |
| chr4_-_120550146 | 0.22 |
ENST00000354960.3 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
| chr2_+_97426631 | 0.22 |
ENST00000377075.2 |
CNNM4 |
cyclin M4 |
| chr4_-_7069760 | 0.22 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr5_+_218356 | 0.21 |
ENST00000264932.6 ENST00000504309.1 ENST00000510361.1 |
SDHA |
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr11_+_128634589 | 0.21 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_145425573 | 0.21 |
ENST00000600064.1 ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr2_-_44223138 | 0.21 |
ENST00000260665.7 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
| chr3_-_167452262 | 0.21 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr12_+_51632666 | 0.21 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr1_-_33502528 | 0.21 |
ENST00000354858.6 |
AK2 |
adenylate kinase 2 |
| chr14_-_106622419 | 0.21 |
ENST00000390604.2 |
IGHV3-16 |
immunoglobulin heavy variable 3-16 (non-functional) |
| chr19_-_15590306 | 0.21 |
ENST00000292609.4 |
PGLYRP2 |
peptidoglycan recognition protein 2 |
| chr3_+_179322573 | 0.21 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_-_33502441 | 0.21 |
ENST00000548033.1 ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2 |
adenylate kinase 2 |
| chr1_+_46152886 | 0.20 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr4_+_41614909 | 0.20 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr16_+_777246 | 0.20 |
ENST00000561546.1 ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr7_-_140624499 | 0.20 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr17_+_13972807 | 0.20 |
ENST00000429152.2 ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10 |
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr4_-_40631859 | 0.20 |
ENST00000295971.7 ENST00000319592.4 |
RBM47 |
RNA binding motif protein 47 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.6 | 5.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 1.7 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.4 | 1.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 1.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.3 | 1.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 1.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.2 | 1.5 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 0.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.7 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.2 | 0.6 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 0.6 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.2 | 0.6 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 2.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 1.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.7 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.4 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 1.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.3 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 5.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.3 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.4 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.2 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.2 | GO:0018013 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.1 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 1.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 1.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.0 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 1.7 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 2.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 1.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 6.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 5.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 1.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 1.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 5.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.3 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 1.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 0.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.5 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 2.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 3.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 5.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 10.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |