Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
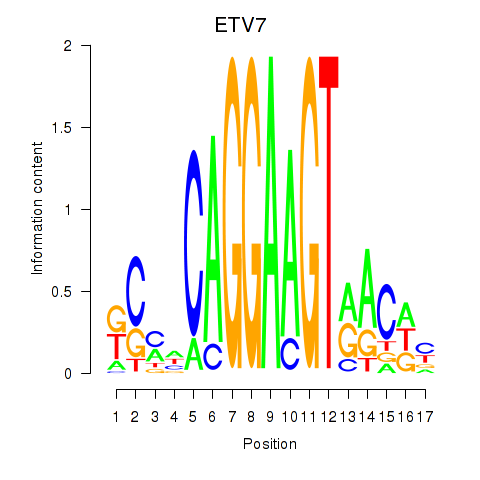
Results for ETV7
Z-value: 0.32
Transcription factors associated with ETV7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV7
|
ENSG00000010030.9 | ETV7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV7 | hg19_v2_chr6_-_36355513_36355578 | 0.65 | 7.9e-02 | Click! |
Activity profile of ETV7 motif
Sorted Z-values of ETV7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_121379739 | 0.39 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr7_-_44365020 | 0.27 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr1_-_209979465 | 0.26 |
ENST00000542854.1 |
IRF6 |
interferon regulatory factor 6 |
| chr16_-_4401258 | 0.24 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr16_-_4401284 | 0.24 |
ENST00000318059.3 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr7_-_44365216 | 0.21 |
ENST00000358707.3 ENST00000457475.1 ENST00000440254.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr1_-_209979375 | 0.21 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr9_-_139891165 | 0.21 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr12_-_52887034 | 0.20 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr16_-_31214051 | 0.20 |
ENST00000350605.4 |
PYCARD |
PYD and CARD domain containing |
| chr8_+_86089460 | 0.19 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr13_+_107029084 | 0.19 |
ENST00000444865.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr11_+_66824276 | 0.18 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr5_-_41510656 | 0.17 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_41510725 | 0.16 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr19_+_17862274 | 0.16 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chr19_+_41222998 | 0.15 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr11_-_118122996 | 0.15 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chrX_+_135618258 | 0.15 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr17_+_78389247 | 0.15 |
ENST00000520136.2 ENST00000520284.1 ENST00000517795.1 ENST00000523228.1 ENST00000523828.1 ENST00000522200.1 ENST00000521565.1 ENST00000518907.1 ENST00000518644.1 ENST00000518901.1 |
ENDOV |
endonuclease V |
| chr12_-_52604607 | 0.15 |
ENST00000551894.1 ENST00000553017.1 |
C12orf80 |
chromosome 12 open reading frame 80 |
| chr8_-_57472154 | 0.15 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr2_-_220436248 | 0.15 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr8_-_28347737 | 0.14 |
ENST00000517673.1 ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16 |
F-box protein 16 |
| chr5_-_35230434 | 0.14 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr3_-_47324242 | 0.14 |
ENST00000456548.1 ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9 |
kinesin family member 9 |
| chrX_-_31285042 | 0.14 |
ENST00000378680.2 ENST00000378723.3 |
DMD |
dystrophin |
| chr15_+_76196234 | 0.13 |
ENST00000540507.1 ENST00000565036.1 ENST00000569054.1 |
FBXO22 |
F-box protein 22 |
| chr11_+_6226782 | 0.13 |
ENST00000316375.2 |
C11orf42 |
chromosome 11 open reading frame 42 |
| chr14_+_67999999 | 0.13 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr12_+_7055767 | 0.13 |
ENST00000447931.2 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr19_+_35739897 | 0.13 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr22_-_36877371 | 0.13 |
ENST00000403313.1 |
TXN2 |
thioredoxin 2 |
| chr22_+_38035459 | 0.13 |
ENST00000357436.4 |
SH3BP1 |
SH3-domain binding protein 1 |
| chr14_-_24804269 | 0.13 |
ENST00000310677.4 ENST00000554068.2 ENST00000559167.1 ENST00000561138.1 |
ADCY4 |
adenylate cyclase 4 |
| chr19_+_35739782 | 0.13 |
ENST00000347609.4 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr10_-_90712520 | 0.13 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr1_-_175161890 | 0.12 |
ENST00000545251.2 ENST00000423313.1 |
KIAA0040 |
KIAA0040 |
| chr16_-_66952742 | 0.12 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr6_+_53659746 | 0.12 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr16_-_66952779 | 0.12 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr17_+_6918064 | 0.12 |
ENST00000546760.1 ENST00000552402.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr8_-_130799134 | 0.12 |
ENST00000276708.4 |
GSDMC |
gasdermin C |
| chr11_+_1861399 | 0.11 |
ENST00000381905.3 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr4_+_74347400 | 0.11 |
ENST00000226355.3 |
AFM |
afamin |
| chr6_-_33267101 | 0.11 |
ENST00000497454.1 |
RGL2 |
ral guanine nucleotide dissociation stimulator-like 2 |
| chr10_+_18948311 | 0.11 |
ENST00000377275.3 |
ARL5B |
ADP-ribosylation factor-like 5B |
| chr3_+_98482175 | 0.11 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr12_+_7055631 | 0.11 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_+_121447469 | 0.11 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr1_+_62439037 | 0.11 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr15_+_26360970 | 0.11 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr9_+_131084846 | 0.11 |
ENST00000608951.1 |
COQ4 |
coenzyme Q4 |
| chr13_+_107028897 | 0.11 |
ENST00000439790.1 ENST00000435024.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr14_-_21567009 | 0.11 |
ENST00000556174.1 ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219 |
zinc finger protein 219 |
| chr1_+_156024525 | 0.11 |
ENST00000368305.4 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr17_+_6918093 | 0.11 |
ENST00000439424.2 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr19_-_291365 | 0.11 |
ENST00000591572.1 ENST00000269812.3 ENST00000434325.2 |
PPAP2C |
phosphatidic acid phosphatase type 2C |
| chr20_-_43883197 | 0.11 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chrX_+_135614293 | 0.11 |
ENST00000370634.3 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr14_+_21387508 | 0.10 |
ENST00000555624.1 |
RP11-84C10.2 |
RP11-84C10.2 |
| chr17_+_6918354 | 0.10 |
ENST00000552775.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr1_+_156024552 | 0.10 |
ENST00000368304.5 ENST00000368302.3 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr1_+_95285896 | 0.10 |
ENST00000446120.2 ENST00000271227.6 ENST00000527077.1 ENST00000529450.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr21_+_18885430 | 0.10 |
ENST00000356275.6 ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr17_+_78388959 | 0.10 |
ENST00000518137.1 ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV |
endonuclease V |
| chr15_+_76196200 | 0.10 |
ENST00000308275.3 ENST00000453211.2 |
FBXO22 |
F-box protein 22 |
| chr10_+_127585093 | 0.10 |
ENST00000368695.1 ENST00000368693.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr3_-_47324008 | 0.10 |
ENST00000425853.1 |
KIF9 |
kinesin family member 9 |
| chr2_+_74685527 | 0.10 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chr5_-_72861484 | 0.10 |
ENST00000296785.3 |
ANKRA2 |
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_40971643 | 0.09 |
ENST00000595483.1 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr3_-_47324079 | 0.09 |
ENST00000352910.4 |
KIF9 |
kinesin family member 9 |
| chr11_-_34535297 | 0.09 |
ENST00000532417.1 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr19_-_41222775 | 0.09 |
ENST00000324464.3 ENST00000450541.1 ENST00000594720.1 |
ADCK4 |
aarF domain containing kinase 4 |
| chr19_+_11651942 | 0.09 |
ENST00000587087.1 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr19_-_40971667 | 0.09 |
ENST00000263368.4 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr2_+_74685413 | 0.09 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr17_-_7155802 | 0.09 |
ENST00000572043.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr16_+_3692931 | 0.09 |
ENST00000407479.1 |
DNASE1 |
deoxyribonuclease I |
| chr11_-_34535332 | 0.09 |
ENST00000257832.2 ENST00000429939.2 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr1_+_156785425 | 0.09 |
ENST00000392302.2 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
| chr17_-_27916555 | 0.09 |
ENST00000394869.3 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr12_+_133195356 | 0.09 |
ENST00000389110.3 ENST00000449132.2 ENST00000343948.4 ENST00000352418.4 ENST00000350048.5 ENST00000351222.4 ENST00000348800.5 ENST00000542301.1 ENST00000536121.1 |
P2RX2 |
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr8_-_82754427 | 0.09 |
ENST00000353788.4 ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16 |
sorting nexin 16 |
| chr2_+_54198210 | 0.09 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr9_-_93925369 | 0.09 |
ENST00000457976.1 |
RP11-305L7.3 |
RP11-305L7.3 |
| chr21_+_18885318 | 0.09 |
ENST00000400166.1 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr20_-_48532046 | 0.09 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr1_+_6086380 | 0.08 |
ENST00000602612.1 ENST00000378087.3 ENST00000341524.1 ENST00000352527.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr2_-_112237835 | 0.08 |
ENST00000442293.1 ENST00000439494.1 |
MIR4435-1HG |
MIR4435-1 host gene (non-protein coding) |
| chr8_-_56685966 | 0.08 |
ENST00000334667.2 |
TMEM68 |
transmembrane protein 68 |
| chr14_-_67878917 | 0.08 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr3_-_47324060 | 0.08 |
ENST00000452770.2 |
KIF9 |
kinesin family member 9 |
| chr7_-_25702669 | 0.08 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr15_-_83474806 | 0.08 |
ENST00000541889.1 ENST00000334574.8 ENST00000561368.1 |
FSD2 |
fibronectin type III and SPRY domain containing 2 |
| chr22_-_36877629 | 0.08 |
ENST00000416967.1 |
TXN2 |
thioredoxin 2 |
| chr10_-_135090338 | 0.08 |
ENST00000415217.3 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr6_-_47009996 | 0.08 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr7_+_1609694 | 0.08 |
ENST00000437621.2 ENST00000457484.2 |
PSMG3-AS1 |
PSMG3 antisense RNA 1 (head to head) |
| chr10_-_135090360 | 0.08 |
ENST00000486609.1 ENST00000445355.3 ENST00000485491.2 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr9_-_37592561 | 0.08 |
ENST00000544379.1 ENST00000377773.5 ENST00000401811.3 ENST00000321301.6 |
TOMM5 |
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr5_-_150466692 | 0.08 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr22_+_32750872 | 0.08 |
ENST00000397468.1 |
RFPL3 |
ret finger protein-like 3 |
| chr6_+_26597155 | 0.08 |
ENST00000274849.1 |
ABT1 |
activator of basal transcription 1 |
| chr14_+_24584508 | 0.08 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr17_-_7835228 | 0.08 |
ENST00000303731.4 ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1 |
trafficking protein particle complex 1 |
| chr17_-_19281203 | 0.08 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr2_+_74056147 | 0.08 |
ENST00000394070.2 ENST00000536064.1 |
STAMBP |
STAM binding protein |
| chr1_-_146644036 | 0.08 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr12_+_123464607 | 0.07 |
ENST00000543566.1 ENST00000315580.5 ENST00000542099.1 ENST00000392435.2 ENST00000413381.2 ENST00000426960.2 ENST00000453766.2 |
ARL6IP4 |
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr5_+_271733 | 0.07 |
ENST00000264933.4 |
PDCD6 |
programmed cell death 6 |
| chr12_-_6772303 | 0.07 |
ENST00000396807.4 ENST00000446105.2 ENST00000341550.4 |
ING4 |
inhibitor of growth family, member 4 |
| chr8_-_100025238 | 0.07 |
ENST00000521696.1 |
RP11-410L14.2 |
RP11-410L14.2 |
| chr1_-_28520384 | 0.07 |
ENST00000305392.3 |
PTAFR |
platelet-activating factor receptor |
| chr9_-_114557207 | 0.07 |
ENST00000374287.3 ENST00000374283.5 |
C9orf84 |
chromosome 9 open reading frame 84 |
| chr1_+_156123318 | 0.07 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr17_-_72542278 | 0.07 |
ENST00000330793.1 |
CD300C |
CD300c molecule |
| chrX_+_152907913 | 0.07 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr2_+_87808725 | 0.07 |
ENST00000413202.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr3_-_11610255 | 0.07 |
ENST00000424529.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr10_+_134150835 | 0.07 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr1_-_249120832 | 0.07 |
ENST00000366472.5 |
SH3BP5L |
SH3-binding domain protein 5-like |
| chr1_-_115323245 | 0.07 |
ENST00000060969.5 ENST00000369528.5 |
SIKE1 |
suppressor of IKBKE 1 |
| chr22_-_31688381 | 0.07 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr22_-_30695471 | 0.07 |
ENST00000434291.1 |
RP1-130H16.18 |
Uncharacterized protein |
| chr19_-_50370799 | 0.07 |
ENST00000600910.1 ENST00000322344.3 ENST00000600573.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr19_+_34287174 | 0.07 |
ENST00000587559.1 ENST00000588637.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr11_-_64084959 | 0.07 |
ENST00000535750.1 ENST00000535126.1 ENST00000539854.1 ENST00000308774.2 |
TRMT112 |
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr1_-_222763214 | 0.07 |
ENST00000350027.4 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr14_-_21270561 | 0.07 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_-_111852128 | 0.07 |
ENST00000308910.4 |
GCSAM |
germinal center-associated, signaling and motility |
| chr1_-_222763240 | 0.07 |
ENST00000352967.4 ENST00000391882.1 ENST00000543857.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr3_-_197282821 | 0.07 |
ENST00000445160.2 ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr14_-_105531759 | 0.07 |
ENST00000329797.3 ENST00000539291.2 ENST00000392585.2 |
GPR132 |
G protein-coupled receptor 132 |
| chr1_-_222763101 | 0.07 |
ENST00000391883.2 ENST00000366890.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr1_-_153538011 | 0.07 |
ENST00000368707.4 |
S100A2 |
S100 calcium binding protein A2 |
| chr6_+_122793058 | 0.07 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr1_-_153538292 | 0.07 |
ENST00000497140.1 ENST00000368708.3 |
S100A2 |
S100 calcium binding protein A2 |
| chr17_+_10600894 | 0.07 |
ENST00000379774.4 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr7_-_100888313 | 0.07 |
ENST00000442303.1 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr14_-_106478603 | 0.06 |
ENST00000390596.2 |
IGHV4-4 |
immunoglobulin heavy variable 4-4 |
| chr4_-_74486217 | 0.06 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_+_87769459 | 0.06 |
ENST00000414030.1 ENST00000437561.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr11_+_126275477 | 0.06 |
ENST00000526727.1 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr4_-_100484825 | 0.06 |
ENST00000273962.3 ENST00000514547.1 ENST00000455368.2 |
TRMT10A |
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr8_+_38243821 | 0.06 |
ENST00000519476.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr13_+_80055284 | 0.06 |
ENST00000218652.7 |
NDFIP2 |
Nedd4 family interacting protein 2 |
| chr4_+_2813946 | 0.06 |
ENST00000442312.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr11_-_64085533 | 0.06 |
ENST00000544844.1 |
TRMT112 |
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr15_-_101835414 | 0.06 |
ENST00000254193.6 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr10_+_88718397 | 0.06 |
ENST00000372017.3 |
SNCG |
synuclein, gamma (breast cancer-specific protein 1) |
| chr7_-_100888337 | 0.06 |
ENST00000223136.4 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr4_-_153456153 | 0.06 |
ENST00000603548.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr16_-_20367584 | 0.06 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chr20_-_2489542 | 0.06 |
ENST00000421216.1 ENST00000381253.1 |
ZNF343 |
zinc finger protein 343 |
| chr7_-_100895414 | 0.06 |
ENST00000435848.1 ENST00000474120.1 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr15_-_101835110 | 0.06 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr3_+_52812523 | 0.06 |
ENST00000540715.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr8_+_48920960 | 0.06 |
ENST00000523111.2 ENST00000523432.1 ENST00000521346.1 ENST00000517630.1 |
UBE2V2 |
ubiquitin-conjugating enzyme E2 variant 2 |
| chr11_-_805224 | 0.06 |
ENST00000347755.5 |
PIDD |
p53-induced death domain protein |
| chr3_+_100120441 | 0.06 |
ENST00000489752.1 |
LNP1 |
leukemia NUP98 fusion partner 1 |
| chr16_+_23569021 | 0.06 |
ENST00000567212.1 ENST00000567264.1 |
UBFD1 |
ubiquitin family domain containing 1 |
| chr21_-_46340770 | 0.06 |
ENST00000397854.3 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr13_+_80055581 | 0.06 |
ENST00000487865.1 |
NDFIP2 |
Nedd4 family interacting protein 2 |
| chr11_-_5531215 | 0.06 |
ENST00000311659.4 |
UBQLN3 |
ubiquilin 3 |
| chr3_+_179322573 | 0.06 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr2_-_235405168 | 0.06 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr17_-_27916621 | 0.06 |
ENST00000225394.3 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr17_+_80416050 | 0.06 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr4_-_170924888 | 0.06 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr11_-_67210930 | 0.06 |
ENST00000453768.2 ENST00000545016.1 ENST00000341356.5 |
CORO1B |
coronin, actin binding protein, 1B |
| chr5_+_271752 | 0.06 |
ENST00000505221.1 ENST00000509581.1 ENST00000507528.1 |
PDCD6 |
programmed cell death 6 |
| chrX_-_7895755 | 0.06 |
ENST00000444736.1 ENST00000537427.1 ENST00000442940.1 |
PNPLA4 |
patatin-like phospholipase domain containing 4 |
| chr11_+_65337901 | 0.06 |
ENST00000309328.3 ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1 |
Sjogren syndrome/scleroderma autoantigen 1 |
| chr14_+_74417192 | 0.06 |
ENST00000554320.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr8_+_145133493 | 0.06 |
ENST00000316052.5 ENST00000525936.1 |
EXOSC4 |
exosome component 4 |
| chr6_+_35704804 | 0.06 |
ENST00000373869.3 |
ARMC12 |
armadillo repeat containing 12 |
| chr8_-_30515693 | 0.06 |
ENST00000355904.4 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr3_+_32993065 | 0.06 |
ENST00000330953.5 |
CCR4 |
chemokine (C-C motif) receptor 4 |
| chr12_-_6483969 | 0.06 |
ENST00000396966.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr12_+_109273806 | 0.06 |
ENST00000228476.3 ENST00000547768.1 |
DAO |
D-amino-acid oxidase |
| chr11_-_71823796 | 0.06 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr11_-_71823715 | 0.06 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr9_-_127952032 | 0.06 |
ENST00000456642.1 ENST00000373546.3 ENST00000373547.4 |
PPP6C |
protein phosphatase 6, catalytic subunit |
| chr4_-_135122903 | 0.06 |
ENST00000421491.3 ENST00000529122.2 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
| chr1_-_152552980 | 0.06 |
ENST00000368787.3 |
LCE3D |
late cornified envelope 3D |
| chr9_-_127952187 | 0.06 |
ENST00000451402.1 ENST00000415905.1 |
PPP6C |
protein phosphatase 6, catalytic subunit |
| chr2_+_238969530 | 0.06 |
ENST00000254663.6 ENST00000555827.1 ENST00000373332.3 ENST00000413463.1 ENST00000409736.2 ENST00000422984.2 ENST00000412508.1 ENST00000429612.2 |
SCLY |
selenocysteine lyase |
| chr2_+_70121075 | 0.06 |
ENST00000409116.1 |
SNRNP27 |
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr14_-_23504337 | 0.06 |
ENST00000361611.6 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr12_-_50616382 | 0.06 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr1_-_153513170 | 0.06 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr19_+_3762645 | 0.05 |
ENST00000330133.4 |
MRPL54 |
mitochondrial ribosomal protein L54 |
| chr20_+_1246908 | 0.05 |
ENST00000381873.3 ENST00000381867.1 |
SNPH |
syntaphilin |
| chr16_-_66968055 | 0.05 |
ENST00000568572.1 |
FAM96B |
family with sequence similarity 96, member B |
| chr14_-_23504432 | 0.05 |
ENST00000425762.2 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr2_-_71062938 | 0.05 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr11_+_122753391 | 0.05 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
| chr12_-_50616122 | 0.05 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.2 | GO:2000412 | activation of MAPK activity involved in innate immune response(GO:0035419) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:1904300 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.0 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.0 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.0 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.0 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0034227 | tRNA thio-modification(GO:0034227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0071065 | dense core granule membrane(GO:0032127) alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.0 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.0 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.0 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |