Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for EZH2
Z-value: 0.41
Transcription factors associated with EZH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EZH2
|
ENSG00000106462.6 | EZH2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EZH2 | hg19_v2_chr7_-_148580563_148580601 | -0.20 | 6.3e-01 | Click! |
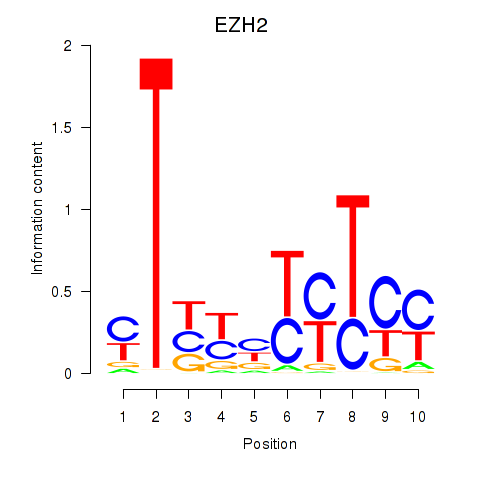
Activity profile of EZH2 motif
Sorted Z-values of EZH2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EZH2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 0.87 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr19_-_51456344 | 0.86 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr10_-_105845674 | 0.86 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr7_+_69064300 | 0.79 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr1_+_86889769 | 0.77 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr15_+_43985084 | 0.68 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr12_-_6483969 | 0.61 |
ENST00000396966.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr12_-_6484376 | 0.59 |
ENST00000360168.3 ENST00000358945.3 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr7_-_41740181 | 0.57 |
ENST00000442711.1 |
INHBA |
inhibin, beta A |
| chr1_-_153588765 | 0.57 |
ENST00000368701.1 ENST00000344616.2 |
S100A14 |
S100 calcium binding protein A14 |
| chr19_-_36004543 | 0.53 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr19_-_36001286 | 0.53 |
ENST00000602679.1 ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN |
dermokine |
| chr8_-_41166953 | 0.52 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr1_-_209824643 | 0.52 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr15_+_43886057 | 0.52 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr19_-_35992780 | 0.49 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr7_-_22233442 | 0.48 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_-_153588334 | 0.48 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr13_-_20805109 | 0.41 |
ENST00000241124.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr12_-_6484715 | 0.39 |
ENST00000228916.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr19_+_35606692 | 0.39 |
ENST00000406242.3 ENST00000454903.2 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr15_+_43985725 | 0.35 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr18_+_29027696 | 0.33 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr19_-_51522955 | 0.33 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr2_-_31637560 | 0.33 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr2_-_113594279 | 0.33 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr19_-_51456321 | 0.32 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr8_+_102504651 | 0.32 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chrX_-_153141302 | 0.31 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chr17_+_55183261 | 0.29 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr7_+_145813453 | 0.29 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr1_+_2036149 | 0.28 |
ENST00000482686.1 ENST00000400920.1 ENST00000486681.1 |
PRKCZ |
protein kinase C, zeta |
| chr3_+_189507432 | 0.27 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr1_+_233749739 | 0.27 |
ENST00000366621.3 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr18_+_47088401 | 0.27 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr11_-_119993734 | 0.26 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_+_109792641 | 0.26 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr12_-_8815299 | 0.26 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr11_-_66496430 | 0.26 |
ENST00000533211.1 |
SPTBN2 |
spectrin, beta, non-erythrocytic 2 |
| chr11_-_5323226 | 0.26 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr4_+_81118647 | 0.26 |
ENST00000415738.2 |
PRDM8 |
PR domain containing 8 |
| chr6_-_136847610 | 0.26 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr2_-_216003127 | 0.26 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr14_-_54423529 | 0.25 |
ENST00000245451.4 ENST00000559087.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr11_+_128563652 | 0.25 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_24797085 | 0.24 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr1_+_2004901 | 0.24 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr5_+_145317356 | 0.24 |
ENST00000511217.1 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr5_+_167181917 | 0.24 |
ENST00000519204.1 |
TENM2 |
teneurin transmembrane protein 2 |
| chr12_-_53343602 | 0.23 |
ENST00000546897.1 ENST00000552551.1 |
KRT8 |
keratin 8 |
| chr14_+_22580233 | 0.23 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr1_+_2005425 | 0.23 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr3_-_197300194 | 0.23 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_+_220325441 | 0.22 |
ENST00000396688.1 |
SPEG |
SPEG complex locus |
| chr10_-_116444371 | 0.22 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr12_-_28125638 | 0.22 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr19_-_51504852 | 0.22 |
ENST00000391806.2 ENST00000347619.4 ENST00000291726.7 ENST00000320838.5 |
KLK8 |
kallikrein-related peptidase 8 |
| chr6_-_31550192 | 0.21 |
ENST00000429299.2 ENST00000446745.2 |
LTB |
lymphotoxin beta (TNF superfamily, member 3) |
| chr16_-_31147020 | 0.21 |
ENST00000568261.1 ENST00000567797.1 ENST00000317508.6 |
PRSS8 |
protease, serine, 8 |
| chr19_+_35607166 | 0.21 |
ENST00000604255.1 ENST00000346446.5 ENST00000344013.6 ENST00000603449.1 ENST00000406988.1 ENST00000605550.1 ENST00000604804.1 ENST00000605552.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr8_+_56792355 | 0.20 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr14_-_99737565 | 0.20 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr8_+_56792377 | 0.20 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chrX_-_151619746 | 0.20 |
ENST00000370314.4 |
GABRA3 |
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr19_-_4338783 | 0.20 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr1_+_209859510 | 0.20 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_+_111717511 | 0.20 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr1_+_95286151 | 0.20 |
ENST00000467909.1 ENST00000422520.2 ENST00000532427.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr1_+_152881014 | 0.20 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr2_+_45168875 | 0.20 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr7_+_86273700 | 0.19 |
ENST00000546348.1 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr5_-_60140009 | 0.19 |
ENST00000505959.1 |
ELOVL7 |
ELOVL fatty acid elongase 7 |
| chr14_-_67859422 | 0.19 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr2_-_165424973 | 0.19 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr12_+_83080659 | 0.19 |
ENST00000321196.3 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_-_147245484 | 0.19 |
ENST00000271348.2 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr12_+_101188547 | 0.18 |
ENST00000546991.1 ENST00000392979.3 |
ANO4 |
anoctamin 4 |
| chr6_+_125540951 | 0.18 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_-_47010061 | 0.18 |
ENST00000371253.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr11_-_44331679 | 0.18 |
ENST00000329255.3 |
ALX4 |
ALX homeobox 4 |
| chr20_-_42815733 | 0.18 |
ENST00000342272.3 |
JPH2 |
junctophilin 2 |
| chr12_-_24102576 | 0.18 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr12_+_66218212 | 0.18 |
ENST00000393578.3 ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2 |
high mobility group AT-hook 2 |
| chr2_+_102608306 | 0.18 |
ENST00000332549.3 |
IL1R2 |
interleukin 1 receptor, type II |
| chr6_-_41130841 | 0.18 |
ENST00000373122.4 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr5_-_141249154 | 0.18 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr17_-_56494882 | 0.17 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr6_-_136847099 | 0.17 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr17_-_56494908 | 0.17 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494713 | 0.17 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr14_-_99737822 | 0.17 |
ENST00000345514.2 ENST00000443726.2 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chrX_-_100662881 | 0.17 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr3_+_111630451 | 0.17 |
ENST00000495180.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_193853927 | 0.17 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr14_-_23623577 | 0.17 |
ENST00000422941.2 ENST00000453702.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr6_+_25279651 | 0.17 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr6_+_53659746 | 0.16 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr6_+_13925318 | 0.16 |
ENST00000423553.2 ENST00000537388.1 |
RNF182 |
ring finger protein 182 |
| chr4_-_80994210 | 0.16 |
ENST00000403729.2 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr5_+_140345820 | 0.16 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr12_+_101188718 | 0.16 |
ENST00000299222.9 ENST00000392977.3 |
ANO4 |
anoctamin 4 |
| chr11_-_124311054 | 0.16 |
ENST00000328064.2 |
OR8B8 |
olfactory receptor, family 8, subfamily B, member 8 |
| chr8_-_127570603 | 0.16 |
ENST00000304916.3 |
FAM84B |
family with sequence similarity 84, member B |
| chr15_-_88799948 | 0.15 |
ENST00000394480.2 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr9_+_133971863 | 0.15 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr11_-_5462744 | 0.15 |
ENST00000380211.1 |
OR51I1 |
olfactory receptor, family 51, subfamily I, member 1 |
| chr9_+_133971909 | 0.15 |
ENST00000247291.3 ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L |
allograft inflammatory factor 1-like |
| chr1_-_116311402 | 0.15 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr9_-_107690420 | 0.14 |
ENST00000423487.2 ENST00000374733.1 ENST00000374736.3 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr8_+_128748308 | 0.14 |
ENST00000377970.2 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
| chr18_-_47721447 | 0.14 |
ENST00000285039.7 |
MYO5B |
myosin VB |
| chr5_-_35230434 | 0.14 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr12_+_20848282 | 0.14 |
ENST00000545604.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_20848377 | 0.14 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr12_-_71031220 | 0.14 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr3_+_111717600 | 0.14 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr19_+_51728316 | 0.14 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
| chrX_-_152989798 | 0.14 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr9_-_124989804 | 0.14 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr1_+_167599330 | 0.14 |
ENST00000367854.3 ENST00000361496.3 |
RCSD1 |
RCSD domain containing 1 |
| chr15_+_90544532 | 0.14 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chrX_-_24665208 | 0.14 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr19_-_54676884 | 0.14 |
ENST00000376591.4 |
TMC4 |
transmembrane channel-like 4 |
| chr11_-_74442430 | 0.13 |
ENST00000376332.3 |
CHRDL2 |
chordin-like 2 |
| chr13_-_108518986 | 0.13 |
ENST00000375915.2 |
FAM155A |
family with sequence similarity 155, member A |
| chr7_-_121944491 | 0.13 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr4_+_84457250 | 0.13 |
ENST00000395226.2 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr5_-_41510656 | 0.13 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_+_113376157 | 0.13 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr4_+_84457529 | 0.13 |
ENST00000264409.4 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr7_+_116660246 | 0.13 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr7_+_20370746 | 0.13 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr17_-_46724186 | 0.13 |
ENST00000433510.2 |
RP11-357H14.17 |
RP11-357H14.17 |
| chr6_-_56507586 | 0.13 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr9_+_12693336 | 0.13 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr10_+_75670862 | 0.13 |
ENST00000446342.1 ENST00000372764.3 ENST00000372762.4 |
PLAU |
plasminogen activator, urokinase |
| chr3_-_124839648 | 0.12 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr17_+_7533439 | 0.12 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr12_-_71031185 | 0.12 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr17_+_7387677 | 0.12 |
ENST00000322644.6 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr4_-_80994619 | 0.12 |
ENST00000404191.1 |
ANTXR2 |
anthrax toxin receptor 2 |
| chrX_+_17653413 | 0.12 |
ENST00000398097.3 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr20_-_55100981 | 0.12 |
ENST00000243913.4 |
GCNT7 |
glucosaminyl (N-acetyl) transferase family member 7 |
| chr16_+_66914264 | 0.12 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr3_+_111718036 | 0.12 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chrX_+_105969893 | 0.12 |
ENST00000255499.2 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_-_47009996 | 0.12 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr3_-_48229846 | 0.11 |
ENST00000302506.3 ENST00000351231.3 ENST00000437972.1 |
CDC25A |
cell division cycle 25A |
| chr10_+_96698406 | 0.11 |
ENST00000260682.6 |
CYP2C9 |
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr14_+_23305760 | 0.11 |
ENST00000311852.6 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
| chr1_-_177134024 | 0.11 |
ENST00000367654.3 |
ASTN1 |
astrotactin 1 |
| chr20_-_62284766 | 0.11 |
ENST00000370053.1 |
STMN3 |
stathmin-like 3 |
| chr4_-_40517984 | 0.11 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr6_-_116381918 | 0.11 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr4_+_155484155 | 0.11 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr3_+_189507523 | 0.11 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr4_+_159443024 | 0.11 |
ENST00000448688.2 |
RXFP1 |
relaxin/insulin-like family peptide receptor 1 |
| chr5_-_141257954 | 0.11 |
ENST00000456271.1 ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1 |
protocadherin 1 |
| chr8_+_128748466 | 0.11 |
ENST00000524013.1 ENST00000520751.1 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
| chr19_-_43702231 | 0.11 |
ENST00000597374.1 ENST00000599371.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr14_+_93118813 | 0.11 |
ENST00000556418.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr14_+_22984601 | 0.11 |
ENST00000390509.1 |
TRAJ28 |
T cell receptor alpha joining 28 |
| chr20_-_23066953 | 0.11 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr19_-_4338838 | 0.10 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr9_-_34372830 | 0.10 |
ENST00000379142.3 |
KIAA1161 |
KIAA1161 |
| chrX_+_135730373 | 0.10 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr5_-_41510725 | 0.10 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chrX_+_135730297 | 0.10 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr11_+_6226782 | 0.10 |
ENST00000316375.2 |
C11orf42 |
chromosome 11 open reading frame 42 |
| chr1_+_117544366 | 0.10 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr7_-_100881109 | 0.10 |
ENST00000308344.5 |
CLDN15 |
claudin 15 |
| chr7_+_20370300 | 0.10 |
ENST00000537992.1 |
ITGB8 |
integrin, beta 8 |
| chr1_+_172389821 | 0.10 |
ENST00000367727.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr1_-_11024258 | 0.10 |
ENST00000418570.2 |
C1orf127 |
chromosome 1 open reading frame 127 |
| chr8_+_21916680 | 0.10 |
ENST00000358242.3 ENST00000415253.1 |
DMTN |
dematin actin binding protein |
| chr11_+_55594695 | 0.10 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr18_-_28742813 | 0.10 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr17_-_38938786 | 0.10 |
ENST00000301656.3 |
KRT27 |
keratin 27 |
| chr3_+_109128837 | 0.10 |
ENST00000497996.1 |
RP11-702L6.4 |
RP11-702L6.4 |
| chr13_-_72441315 | 0.10 |
ENST00000305425.4 ENST00000313174.7 ENST00000354591.4 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr15_-_68498376 | 0.10 |
ENST00000540479.1 ENST00000395465.3 |
CALML4 |
calmodulin-like 4 |
| chr2_+_90108504 | 0.10 |
ENST00000390271.2 |
IGKV6D-41 |
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr1_+_214161854 | 0.10 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr5_-_24645078 | 0.10 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr1_-_177133818 | 0.09 |
ENST00000424564.2 ENST00000361833.2 |
ASTN1 |
astrotactin 1 |
| chr11_+_65554493 | 0.09 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr9_-_117568365 | 0.09 |
ENST00000374045.4 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
| chr8_+_21916710 | 0.09 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chrX_-_19002696 | 0.09 |
ENST00000379942.4 |
PHKA2 |
phosphorylase kinase, alpha 2 (liver) |
| chr1_+_172422026 | 0.09 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_-_205391178 | 0.09 |
ENST00000367153.4 ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1 |
LEM domain containing 1 |
| chr6_+_44194762 | 0.09 |
ENST00000371708.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_-_150884332 | 0.09 |
ENST00000275838.1 ENST00000422024.1 ENST00000434669.1 |
ASB10 |
ankyrin repeat and SOCS box containing 10 |
| chr14_+_60975644 | 0.09 |
ENST00000327720.5 |
SIX6 |
SIX homeobox 6 |
| chr10_+_71389983 | 0.09 |
ENST00000373279.4 |
C10orf35 |
chromosome 10 open reading frame 35 |
| chrX_-_24665353 | 0.09 |
ENST00000379144.2 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr8_+_99956759 | 0.09 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr12_+_175930 | 0.09 |
ENST00000538872.1 ENST00000326261.4 |
IQSEC3 |
IQ motif and Sec7 domain 3 |
| chr2_+_220462560 | 0.09 |
ENST00000456909.1 ENST00000295641.10 |
STK11IP |
serine/threonine kinase 11 interacting protein |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 1.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 1.2 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.4 | GO:0070668 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.4 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.8 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.3 | GO:0072192 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.3 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.2 | GO:0098905 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.2 | GO:2000974 | inhibition of neuroepithelial cell differentiation(GO:0002085) trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.0 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 1.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.3 | GO:0061589 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.3 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 1.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.0 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:0031960 | response to corticosteroid(GO:0031960) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.0 | 0.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.0 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0010820 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.0 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0035054 | septum secundum development(GO:0003285) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:2000777 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.0 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.0 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |