Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
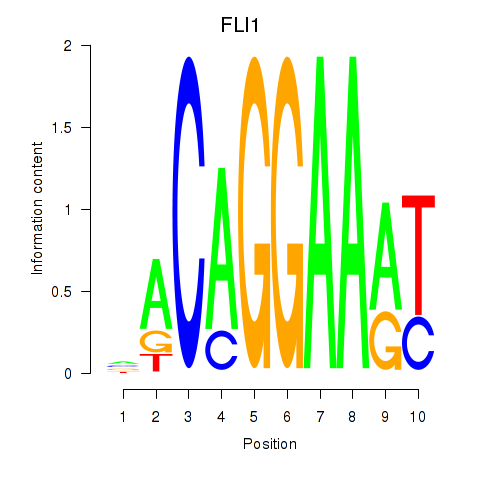
Results for FLI1
Z-value: 0.71
Transcription factors associated with FLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FLI1
|
ENSG00000151702.12 | FLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FLI1 | hg19_v2_chr11_+_128563652_128563689, hg19_v2_chr11_+_128634589_128634685, hg19_v2_chr11_+_128563948_128564003 | 0.44 | 2.7e-01 | Click! |
Activity profile of FLI1 motif
Sorted Z-values of FLI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FLI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 1.46 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 1.36 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43985725 | 1.34 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr13_-_20805109 | 1.06 |
ENST00000241124.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr3_-_121379739 | 0.99 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr1_+_86889769 | 0.80 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr3_-_151034734 | 0.80 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr6_+_125474795 | 0.73 |
ENST00000304877.13 ENST00000534000.1 ENST00000368402.5 ENST00000368388.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr4_+_40198527 | 0.69 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr1_-_201368653 | 0.68 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr6_+_125474992 | 0.66 |
ENST00000528193.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_-_201368707 | 0.65 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr6_+_125474939 | 0.65 |
ENST00000527711.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_+_125475335 | 0.64 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr11_+_18154059 | 0.58 |
ENST00000531264.1 |
MRGPRX3 |
MAS-related GPR, member X3 |
| chr19_+_35739897 | 0.53 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr11_-_72492903 | 0.52 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr20_+_58296265 | 0.51 |
ENST00000395636.2 ENST00000361300.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr19_+_35739782 | 0.50 |
ENST00000347609.4 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr1_-_175161890 | 0.50 |
ENST00000545251.2 ENST00000423313.1 |
KIAA0040 |
KIAA0040 |
| chr4_-_10023095 | 0.49 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr1_+_153330322 | 0.48 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr18_-_74844713 | 0.47 |
ENST00000397860.3 |
MBP |
myelin basic protein |
| chr1_+_35220613 | 0.46 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr1_+_6086354 | 0.46 |
ENST00000389632.4 ENST00000428161.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr12_-_54867352 | 0.45 |
ENST00000305879.5 |
GTSF1 |
gametocyte specific factor 1 |
| chr17_-_39677971 | 0.44 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr19_-_6767431 | 0.44 |
ENST00000437152.3 ENST00000597687.1 |
SH2D3A |
SH2 domain containing 3A |
| chrX_+_102469997 | 0.42 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr18_+_61420169 | 0.41 |
ENST00000425392.1 ENST00000336429.2 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_-_6740802 | 0.40 |
ENST00000431922.1 |
LPAR5 |
lysophosphatidic acid receptor 5 |
| chr8_+_22438009 | 0.40 |
ENST00000409417.1 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr16_+_57653854 | 0.38 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57653989 | 0.38 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr6_+_28048753 | 0.37 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr8_+_145133493 | 0.37 |
ENST00000316052.5 ENST00000525936.1 |
EXOSC4 |
exosome component 4 |
| chr14_+_65171315 | 0.37 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr20_-_43883197 | 0.37 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr2_+_18059906 | 0.37 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr12_+_107712173 | 0.37 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr4_-_90759440 | 0.36 |
ENST00000336904.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr22_+_45098067 | 0.36 |
ENST00000336985.6 ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5 PRR5-ARHGAP8 |
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr6_-_32784687 | 0.35 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr10_-_98031265 | 0.35 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr19_+_41281416 | 0.35 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr1_-_175162048 | 0.34 |
ENST00000444639.1 |
KIAA0040 |
KIAA0040 |
| chr6_-_33267101 | 0.34 |
ENST00000497454.1 |
RGL2 |
ral guanine nucleotide dissociation stimulator-like 2 |
| chr19_+_35739280 | 0.34 |
ENST00000602122.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr20_+_58630972 | 0.34 |
ENST00000313426.1 |
C20orf197 |
chromosome 20 open reading frame 197 |
| chr3_-_141747950 | 0.33 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_39928106 | 0.33 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr7_+_150811705 | 0.33 |
ENST00000335367.3 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_153044083 | 0.33 |
ENST00000341611.2 |
SPRR2B |
small proline-rich protein 2B |
| chr10_-_98031310 | 0.32 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr4_+_20702059 | 0.32 |
ENST00000444671.2 ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL |
PARK2 co-regulated-like |
| chr12_+_53342625 | 0.31 |
ENST00000388837.2 ENST00000550600.1 ENST00000388835.3 |
KRT18 |
keratin 18 |
| chr19_-_291133 | 0.31 |
ENST00000327790.3 |
PPAP2C |
phosphatidic acid phosphatase type 2C |
| chr19_+_41281282 | 0.31 |
ENST00000263369.3 |
MIA |
melanoma inhibitory activity |
| chr19_-_19739007 | 0.30 |
ENST00000586703.1 ENST00000591042.1 ENST00000407877.3 |
LPAR2 |
lysophosphatidic acid receptor 2 |
| chr5_-_35230434 | 0.30 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr14_+_21510385 | 0.30 |
ENST00000298690.4 |
RNASE7 |
ribonuclease, RNase A family, 7 |
| chr1_-_156786634 | 0.30 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr1_-_156786530 | 0.30 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chrX_-_128788914 | 0.29 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr19_+_41281060 | 0.29 |
ENST00000594436.1 ENST00000597784.1 |
MIA |
melanoma inhibitory activity |
| chr1_+_144339738 | 0.29 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr20_+_35201993 | 0.29 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr11_+_59522900 | 0.29 |
ENST00000529177.1 |
STX3 |
syntaxin 3 |
| chr17_+_78389247 | 0.29 |
ENST00000520136.2 ENST00000520284.1 ENST00000517795.1 ENST00000523228.1 ENST00000523828.1 ENST00000522200.1 ENST00000521565.1 ENST00000518907.1 ENST00000518644.1 ENST00000518901.1 |
ENDOV |
endonuclease V |
| chr17_-_7493390 | 0.28 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr19_+_56186557 | 0.28 |
ENST00000270460.6 |
EPN1 |
epsin 1 |
| chr14_+_68286478 | 0.28 |
ENST00000487861.1 |
RAD51B |
RAD51 paralog B |
| chr19_-_11688447 | 0.28 |
ENST00000590420.1 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr20_-_1306391 | 0.28 |
ENST00000339987.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr11_+_47270436 | 0.28 |
ENST00000395397.3 ENST00000405576.1 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr11_+_47270475 | 0.28 |
ENST00000481889.2 ENST00000436778.1 ENST00000531660.1 ENST00000407404.1 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr1_-_183560011 | 0.28 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr8_-_42397037 | 0.28 |
ENST00000342228.3 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
| chr14_+_67999999 | 0.28 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr6_-_42419649 | 0.28 |
ENST00000372922.4 ENST00000541110.1 ENST00000372917.4 |
TRERF1 |
transcriptional regulating factor 1 |
| chr6_-_41130914 | 0.27 |
ENST00000373113.3 ENST00000338469.3 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr19_-_291365 | 0.27 |
ENST00000591572.1 ENST00000269812.3 ENST00000434325.2 |
PPAP2C |
phosphatidic acid phosphatase type 2C |
| chr11_+_59522837 | 0.27 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr17_-_76123101 | 0.27 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
| chr17_-_76124812 | 0.27 |
ENST00000592063.1 ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6 |
transmembrane channel-like 6 |
| chr2_+_233925064 | 0.27 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr17_+_48503519 | 0.26 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr1_-_183559693 | 0.26 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr17_+_48503603 | 0.26 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr6_+_7541808 | 0.26 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr9_-_104249400 | 0.26 |
ENST00000374848.3 |
TMEM246 |
transmembrane protein 246 |
| chr6_+_41606176 | 0.26 |
ENST00000441667.1 ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI |
MyoD family inhibitor |
| chr1_-_68516393 | 0.25 |
ENST00000395201.1 |
DIRAS3 |
DIRAS family, GTP-binding RAS-like 3 |
| chr17_+_54671047 | 0.25 |
ENST00000332822.4 |
NOG |
noggin |
| chr1_+_82266053 | 0.25 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr9_-_117150243 | 0.25 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr16_+_50300427 | 0.25 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr2_-_229046361 | 0.25 |
ENST00000392056.3 |
SPHKAP |
SPHK1 interactor, AKAP domain containing |
| chr1_-_153066998 | 0.25 |
ENST00000368750.3 |
SPRR2E |
small proline-rich protein 2E |
| chr1_+_32716840 | 0.25 |
ENST00000336890.5 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr2_-_229046330 | 0.25 |
ENST00000344657.5 |
SPHKAP |
SPHK1 interactor, AKAP domain containing |
| chr4_+_15471489 | 0.25 |
ENST00000424120.1 ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
| chr1_-_27286897 | 0.25 |
ENST00000320567.5 |
C1orf172 |
chromosome 1 open reading frame 172 |
| chr11_-_72433346 | 0.24 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_+_1077393 | 0.24 |
ENST00000590577.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr11_+_394196 | 0.24 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr11_+_36397528 | 0.24 |
ENST00000311599.5 ENST00000378867.3 |
PRR5L |
proline rich 5 like |
| chr4_+_89299885 | 0.24 |
ENST00000380265.5 ENST00000273960.3 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr18_-_52989217 | 0.24 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr17_-_4843316 | 0.24 |
ENST00000544061.2 |
SLC25A11 |
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr11_+_5646213 | 0.23 |
ENST00000429814.2 |
TRIM34 |
tripartite motif containing 34 |
| chr16_-_11680791 | 0.23 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr11_+_59522532 | 0.23 |
ENST00000337979.4 ENST00000535361.1 |
STX3 |
syntaxin 3 |
| chr6_+_18387570 | 0.23 |
ENST00000259939.3 |
RNF144B |
ring finger protein 144B |
| chr2_-_235405168 | 0.23 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chrX_+_105937068 | 0.23 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_-_112408661 | 0.23 |
ENST00000368662.5 |
TUBE1 |
tubulin, epsilon 1 |
| chr4_+_89299994 | 0.23 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chrX_+_135614293 | 0.23 |
ENST00000370634.3 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr9_-_104249319 | 0.23 |
ENST00000374847.1 |
TMEM246 |
transmembrane protein 246 |
| chr2_+_103035102 | 0.23 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr5_+_35856951 | 0.23 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr18_-_47376197 | 0.23 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr11_-_72432950 | 0.23 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_+_4843679 | 0.23 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr1_+_6086380 | 0.23 |
ENST00000602612.1 ENST00000378087.3 ENST00000341524.1 ENST00000352527.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr10_+_112631547 | 0.23 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr17_-_7166500 | 0.23 |
ENST00000575313.1 ENST00000397317.4 |
CLDN7 |
claudin 7 |
| chr9_+_116917807 | 0.22 |
ENST00000356083.3 |
COL27A1 |
collagen, type XXVII, alpha 1 |
| chr13_-_86373536 | 0.22 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr14_-_92333873 | 0.22 |
ENST00000435962.2 |
TC2N |
tandem C2 domains, nuclear |
| chr6_-_111888474 | 0.22 |
ENST00000368735.1 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr1_+_212738676 | 0.22 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr14_+_65171099 | 0.22 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_-_153029980 | 0.22 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr8_-_125577940 | 0.22 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chr8_+_61591337 | 0.22 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chrX_-_40594755 | 0.22 |
ENST00000324817.1 |
MED14 |
mediator complex subunit 14 |
| chr1_+_26869597 | 0.22 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr16_+_2205755 | 0.22 |
ENST00000326181.6 |
TRAF7 |
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr20_+_35201857 | 0.21 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr6_+_42981922 | 0.21 |
ENST00000326974.4 ENST00000244670.8 |
KLHDC3 |
kelch domain containing 3 |
| chr17_-_7167279 | 0.21 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr21_+_18885430 | 0.21 |
ENST00000356275.6 ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr12_+_110172572 | 0.21 |
ENST00000358906.3 |
FAM222A |
family with sequence similarity 222, member A |
| chr8_+_42911552 | 0.21 |
ENST00000525699.1 ENST00000529687.1 |
FNTA |
farnesyltransferase, CAAX box, alpha |
| chr16_+_4845379 | 0.21 |
ENST00000588606.1 ENST00000586005.1 |
SMIM22 |
small integral membrane protein 22 |
| chr3_+_57741957 | 0.21 |
ENST00000295951.3 |
SLMAP |
sarcolemma associated protein |
| chr1_+_229440129 | 0.21 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr2_+_113342011 | 0.21 |
ENST00000324913.5 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr18_-_52989525 | 0.21 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_+_220325977 | 0.21 |
ENST00000396686.1 ENST00000396689.2 |
SPEG |
SPEG complex locus |
| chr1_+_95285896 | 0.21 |
ENST00000446120.2 ENST00000271227.6 ENST00000527077.1 ENST00000529450.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr11_+_6226782 | 0.21 |
ENST00000316375.2 |
C11orf42 |
chromosome 11 open reading frame 42 |
| chr1_-_24513737 | 0.20 |
ENST00000374421.3 ENST00000374418.3 ENST00000327535.1 ENST00000327575.2 |
IFNLR1 |
interferon, lambda receptor 1 |
| chr12_-_22094336 | 0.20 |
ENST00000326684.4 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_-_220436248 | 0.20 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr19_+_56186606 | 0.20 |
ENST00000085079.7 |
EPN1 |
epsin 1 |
| chr2_+_113342163 | 0.20 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_+_17862274 | 0.20 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chr3_-_128840604 | 0.20 |
ENST00000476465.1 ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43 |
RAB43, member RAS oncogene family |
| chr6_+_112408768 | 0.20 |
ENST00000368656.2 ENST00000604268.1 |
FAM229B |
family with sequence similarity 229, member B |
| chr1_-_55230165 | 0.20 |
ENST00000371279.3 |
PARS2 |
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr6_+_26251835 | 0.20 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr8_-_41754231 | 0.20 |
ENST00000265709.8 |
ANK1 |
ankyrin 1, erythrocytic |
| chr1_-_43751230 | 0.20 |
ENST00000523677.1 |
C1orf210 |
chromosome 1 open reading frame 210 |
| chr2_+_233734994 | 0.20 |
ENST00000331342.2 |
C2orf82 |
chromosome 2 open reading frame 82 |
| chrX_-_20236970 | 0.20 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr8_-_144699628 | 0.20 |
ENST00000529048.1 ENST00000529064.1 |
TSTA3 |
tissue specific transplantation antigen P35B |
| chr17_+_78388959 | 0.19 |
ENST00000518137.1 ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV |
endonuclease V |
| chr11_-_93276582 | 0.19 |
ENST00000298966.2 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
| chr18_-_24237339 | 0.19 |
ENST00000580191.1 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chr6_+_3849584 | 0.19 |
ENST00000380274.1 ENST00000380272.3 |
FAM50B |
family with sequence similarity 50, member B |
| chr17_-_7165662 | 0.19 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr20_+_42984330 | 0.19 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr11_-_104034827 | 0.19 |
ENST00000393158.2 |
PDGFD |
platelet derived growth factor D |
| chr6_+_33172407 | 0.19 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr21_+_18885318 | 0.19 |
ENST00000400166.1 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr3_-_112693865 | 0.19 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr8_-_144699668 | 0.19 |
ENST00000425753.2 |
TSTA3 |
tissue specific transplantation antigen P35B |
| chr19_-_54676846 | 0.19 |
ENST00000301187.4 |
TMC4 |
transmembrane channel-like 4 |
| chr3_-_119396193 | 0.18 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr17_+_21191341 | 0.18 |
ENST00000526076.2 ENST00000361818.5 ENST00000316920.6 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
| chr4_-_186733363 | 0.18 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr14_-_100842588 | 0.18 |
ENST00000556645.1 ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS |
tryptophanyl-tRNA synthetase |
| chr15_-_44486632 | 0.18 |
ENST00000484674.1 |
FRMD5 |
FERM domain containing 5 |
| chr8_-_144660771 | 0.18 |
ENST00000449291.2 |
NAPRT1 |
nicotinate phosphoribosyltransferase domain containing 1 |
| chr19_+_16296191 | 0.18 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr4_+_78079570 | 0.18 |
ENST00000509972.1 |
CCNG2 |
cyclin G2 |
| chr8_-_86253888 | 0.18 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr7_+_99195878 | 0.18 |
ENST00000453227.1 ENST00000429679.1 |
GS1-259H13.2 |
GS1-259H13.2 |
| chr16_-_4466565 | 0.18 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr11_-_75017734 | 0.18 |
ENST00000532525.1 |
ARRB1 |
arrestin, beta 1 |
| chr20_+_9494987 | 0.18 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr8_-_144660485 | 0.18 |
ENST00000276844.7 ENST00000340490.3 ENST00000435154.3 ENST00000426292.3 |
NAPRT1 |
nicotinate phosphoribosyltransferase domain containing 1 |
| chr17_+_7461613 | 0.18 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_11688500 | 0.18 |
ENST00000433365.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr1_-_205744205 | 0.18 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr19_+_6887571 | 0.18 |
ENST00000250572.8 ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr8_-_12668962 | 0.18 |
ENST00000534827.1 |
RP11-252C15.1 |
RP11-252C15.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 2.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.6 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.4 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.6 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.6 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.2 | GO:1904760 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.7 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.2 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:2000407 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0061346 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.4 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 1.1 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.0 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.0 | GO:1990592 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 2.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) |
| 0.0 | 0.0 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.0 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.0 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.0 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 0.7 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 1.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.0 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 0.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.6 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.2 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0030291 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.0 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.0 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 4.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |