Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
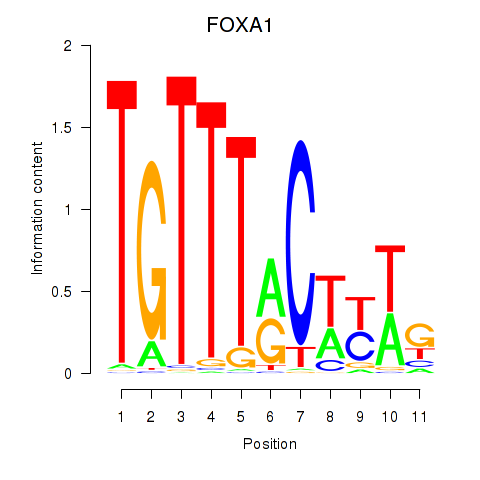
Results for FOXA1
Z-value: 1.02
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | FOXA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | -0.51 | 1.9e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91572278 | 1.62 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr9_-_95186739 | 1.42 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr5_+_156712372 | 0.91 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr5_-_41510725 | 0.69 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_41510656 | 0.69 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_-_67802549 | 0.69 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr18_-_25616519 | 0.64 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr17_-_19651668 | 0.61 |
ENST00000494157.2 ENST00000225740.6 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chrX_+_9431324 | 0.60 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr6_-_117747015 | 0.59 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr10_+_124320156 | 0.58 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr10_+_124320195 | 0.57 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr8_+_70404996 | 0.54 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr13_+_76334498 | 0.52 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr4_+_55095264 | 0.49 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_-_87804815 | 0.49 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr1_+_223101757 | 0.49 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr6_-_56707943 | 0.47 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr2_-_192711968 | 0.46 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr6_+_108977520 | 0.45 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr9_-_20622478 | 0.42 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr5_-_127674883 | 0.41 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr1_-_232651312 | 0.40 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr2_+_172950227 | 0.40 |
ENST00000341900.6 |
DLX1 |
distal-less homeobox 1 |
| chr9_-_89562104 | 0.39 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr3_+_69985734 | 0.38 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_158345462 | 0.38 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr13_+_76334795 | 0.35 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr1_+_101361782 | 0.34 |
ENST00000357650.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_150157444 | 0.33 |
ENST00000526627.1 |
SMIM3 |
small integral membrane protein 3 |
| chr20_-_45984401 | 0.33 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr6_-_88875654 | 0.33 |
ENST00000535130.1 |
CNR1 |
cannabinoid receptor 1 (brain) |
| chr5_+_150157860 | 0.32 |
ENST00000600109.1 |
AC010441.1 |
AC010441.1 |
| chr13_+_76334567 | 0.30 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr9_-_123239632 | 0.30 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chrX_+_85969626 | 0.30 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr17_+_79953310 | 0.30 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chrX_+_22050546 | 0.29 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr10_-_90751038 | 0.28 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr12_-_71551868 | 0.28 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr3_+_148545586 | 0.27 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr15_+_96869165 | 0.27 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_40713573 | 0.26 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chr12_-_71551652 | 0.26 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr8_+_77593448 | 0.26 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr3_-_145878954 | 0.25 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr20_+_31823792 | 0.25 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr8_+_77593474 | 0.25 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr12_-_92536433 | 0.25 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr1_+_198126093 | 0.24 |
ENST00000367385.4 ENST00000442588.1 ENST00000538004.1 |
NEK7 |
NIMA-related kinase 7 |
| chr3_+_141106643 | 0.24 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr3_+_148447887 | 0.24 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr7_+_134430212 | 0.24 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr3_-_71632894 | 0.23 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chr2_-_233877912 | 0.23 |
ENST00000264051.3 |
NGEF |
neuronal guanine nucleotide exchange factor |
| chr7_+_134576317 | 0.23 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr11_-_9286921 | 0.23 |
ENST00000328194.3 |
DENND5A |
DENN/MADD domain containing 5A |
| chr10_+_104535994 | 0.23 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr2_+_169757750 | 0.22 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr11_-_102401469 | 0.22 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr5_+_102201509 | 0.22 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_102201430 | 0.22 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_61547405 | 0.22 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr5_-_137674000 | 0.21 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr3_-_105588231 | 0.21 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_-_182545603 | 0.20 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr8_+_95908041 | 0.20 |
ENST00000396113.1 ENST00000519136.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr1_-_27240455 | 0.20 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr15_+_75335604 | 0.20 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr5_+_147774275 | 0.20 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr11_+_120973375 | 0.19 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr9_+_6413317 | 0.19 |
ENST00000276893.5 ENST00000381373.3 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr18_+_3449330 | 0.18 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr2_+_152266604 | 0.18 |
ENST00000430328.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr2_+_88047606 | 0.18 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr20_+_31595406 | 0.17 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr16_-_4896205 | 0.17 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr7_+_134576151 | 0.17 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr16_-_87970122 | 0.16 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr11_+_35684288 | 0.16 |
ENST00000299413.5 |
TRIM44 |
tripartite motif containing 44 |
| chr18_+_72922710 | 0.15 |
ENST00000322038.5 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
| chr1_+_104159999 | 0.15 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr12_+_103981044 | 0.15 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr12_-_56122761 | 0.15 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr6_-_111804393 | 0.14 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr21_-_43735446 | 0.14 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr16_+_4896659 | 0.14 |
ENST00000592120.1 |
UBN1 |
ubinuclein 1 |
| chr10_-_81320151 | 0.14 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chr3_+_69985792 | 0.14 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr1_-_177939348 | 0.14 |
ENST00000464631.2 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
| chr6_+_74405501 | 0.13 |
ENST00000437994.2 ENST00000422508.2 |
CD109 |
CD109 molecule |
| chr5_-_133510456 | 0.13 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr22_+_35462129 | 0.13 |
ENST00000404699.2 ENST00000308700.6 |
ISX |
intestine-specific homeobox |
| chr6_+_74405804 | 0.13 |
ENST00000287097.5 |
CD109 |
CD109 molecule |
| chr2_-_152146385 | 0.13 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr12_-_120765565 | 0.13 |
ENST00000423423.3 ENST00000308366.4 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr13_+_50589390 | 0.13 |
ENST00000360473.4 ENST00000312942.1 |
KCNRG |
potassium channel regulator |
| chr18_+_66465302 | 0.13 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr12_+_121416437 | 0.13 |
ENST00000402929.1 ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A |
HNF1 homeobox A |
| chr12_-_56123444 | 0.12 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr2_-_3521518 | 0.12 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr11_-_116663127 | 0.12 |
ENST00000433069.1 ENST00000542499.1 |
APOA5 |
apolipoprotein A-V |
| chr4_+_187187098 | 0.12 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chrX_-_47863348 | 0.12 |
ENST00000376943.3 ENST00000396965.1 ENST00000305127.6 |
ZNF182 |
zinc finger protein 182 |
| chr3_+_108321623 | 0.11 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr19_+_50380917 | 0.11 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr19_+_50380682 | 0.11 |
ENST00000221543.5 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr21_+_33671160 | 0.11 |
ENST00000303645.5 |
MRAP |
melanocortin 2 receptor accessory protein |
| chr18_+_70536215 | 0.11 |
ENST00000578967.1 |
RP11-676J15.1 |
RP11-676J15.1 |
| chr16_-_68034470 | 0.11 |
ENST00000412757.2 |
DPEP2 |
dipeptidase 2 |
| chr17_-_40897043 | 0.11 |
ENST00000428826.2 ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1 |
enhancer of zeste homolog 1 (Drosophila) |
| chr19_+_42806250 | 0.11 |
ENST00000598490.1 ENST00000341747.3 |
PRR19 |
proline rich 19 |
| chr1_+_116654376 | 0.11 |
ENST00000369500.3 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
| chr4_+_170581213 | 0.11 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr8_-_116681221 | 0.10 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr3_-_194030493 | 0.10 |
ENST00000456816.1 ENST00000414120.1 ENST00000429578.1 |
LINC00887 |
long intergenic non-protein coding RNA 887 |
| chr10_+_91092241 | 0.10 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr14_+_64971292 | 0.10 |
ENST00000358738.3 ENST00000394712.2 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chrX_+_135730297 | 0.10 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr11_+_57308979 | 0.10 |
ENST00000457912.1 |
SMTNL1 |
smoothelin-like 1 |
| chr5_+_95998746 | 0.10 |
ENST00000508608.1 |
CAST |
calpastatin |
| chr4_-_155533787 | 0.10 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr4_+_165675269 | 0.10 |
ENST00000507311.1 |
RP11-294O2.2 |
RP11-294O2.2 |
| chrX_+_135730373 | 0.10 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr12_+_96588143 | 0.10 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_61547894 | 0.09 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr1_-_57431679 | 0.09 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr4_+_26321284 | 0.09 |
ENST00000506956.1 ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_+_116165754 | 0.09 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr2_+_65283506 | 0.09 |
ENST00000377990.2 |
CEP68 |
centrosomal protein 68kDa |
| chr20_+_44420617 | 0.09 |
ENST00000449078.1 ENST00000456939.1 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr1_-_95391315 | 0.09 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr11_-_124622134 | 0.09 |
ENST00000326621.5 |
VSIG2 |
V-set and immunoglobulin domain containing 2 |
| chr3_+_137728842 | 0.09 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr8_+_95907993 | 0.09 |
ENST00000523378.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_+_65283529 | 0.09 |
ENST00000546106.1 ENST00000537589.1 ENST00000260569.4 |
CEP68 |
centrosomal protein 68kDa |
| chr13_-_31038370 | 0.08 |
ENST00000399489.1 ENST00000339872.4 |
HMGB1 |
high mobility group box 1 |
| chr6_+_84222194 | 0.08 |
ENST00000536636.1 |
PRSS35 |
protease, serine, 35 |
| chr7_-_7782204 | 0.08 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chr3_-_71294304 | 0.08 |
ENST00000498215.1 |
FOXP1 |
forkhead box P1 |
| chr10_+_124739911 | 0.08 |
ENST00000405485.1 |
PSTK |
phosphoseryl-tRNA kinase |
| chr17_+_36873677 | 0.08 |
ENST00000471200.1 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_+_179345173 | 0.08 |
ENST00000234453.5 |
PLEKHA3 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr19_-_15443318 | 0.08 |
ENST00000360016.5 |
BRD4 |
bromodomain containing 4 |
| chr7_+_134832808 | 0.08 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr20_+_44420570 | 0.08 |
ENST00000372622.3 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr2_-_121223697 | 0.08 |
ENST00000593290.1 |
FLJ14816 |
long intergenic non-protein coding RNA 1101 |
| chr19_-_18902106 | 0.08 |
ENST00000542601.2 ENST00000425807.1 ENST00000222271.2 |
COMP |
cartilage oligomeric matrix protein |
| chr14_+_64970662 | 0.08 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr10_+_124739964 | 0.08 |
ENST00000406217.2 |
PSTK |
phosphoseryl-tRNA kinase |
| chr2_-_172017343 | 0.08 |
ENST00000431350.2 ENST00000360843.3 |
TLK1 |
tousled-like kinase 1 |
| chr14_-_24911448 | 0.08 |
ENST00000555355.1 ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chrX_+_129473916 | 0.08 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr13_-_45775162 | 0.07 |
ENST00000405872.1 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
| chr14_-_94789663 | 0.07 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr6_-_90025011 | 0.07 |
ENST00000402938.3 |
GABRR2 |
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr9_-_14910420 | 0.07 |
ENST00000380880.3 |
FREM1 |
FRAS1 related extracellular matrix 1 |
| chr12_-_68647281 | 0.07 |
ENST00000328087.4 ENST00000538666.1 |
IL22 |
interleukin 22 |
| chr6_+_84222220 | 0.07 |
ENST00000369700.3 |
PRSS35 |
protease, serine, 35 |
| chr12_-_772901 | 0.07 |
ENST00000305108.4 |
NINJ2 |
ninjurin 2 |
| chr15_+_71389281 | 0.07 |
ENST00000355327.3 |
THSD4 |
thrombospondin, type I, domain containing 4 |
| chr20_-_43150601 | 0.07 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr20_+_306177 | 0.07 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr10_+_102891048 | 0.07 |
ENST00000467928.2 |
TLX1 |
T-cell leukemia homeobox 1 |
| chr2_+_204801471 | 0.07 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chrX_+_37865804 | 0.07 |
ENST00000297875.2 ENST00000357972.5 |
SYTL5 |
synaptotagmin-like 5 |
| chr1_+_145883868 | 0.07 |
ENST00000447947.2 |
GPR89C |
G protein-coupled receptor 89C |
| chr6_-_90024967 | 0.07 |
ENST00000602399.1 |
GABRR2 |
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr1_+_43855560 | 0.07 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr20_-_50179368 | 0.07 |
ENST00000609943.1 ENST00000609507.1 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr8_-_71316021 | 0.07 |
ENST00000452400.2 |
NCOA2 |
nuclear receptor coactivator 2 |
| chr5_+_161495038 | 0.07 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_-_89927151 | 0.06 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr1_+_35734562 | 0.06 |
ENST00000314607.6 ENST00000373297.2 |
ZMYM4 |
zinc finger, MYM-type 4 |
| chr1_+_74701062 | 0.06 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr1_+_199996733 | 0.06 |
ENST00000236914.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr10_-_32217717 | 0.06 |
ENST00000396144.4 ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12 |
Rho GTPase activating protein 12 |
| chr1_-_43855479 | 0.06 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr10_-_73848086 | 0.06 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_+_80231466 | 0.06 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr7_-_99698338 | 0.06 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr9_+_82187630 | 0.06 |
ENST00000265284.6 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr7_+_28452130 | 0.06 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr6_+_161123270 | 0.06 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr20_+_306221 | 0.06 |
ENST00000342665.2 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr9_+_82186872 | 0.05 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_+_16178317 | 0.05 |
ENST00000344824.6 ENST00000538887.1 |
TPM4 |
tropomyosin 4 |
| chr2_+_150187020 | 0.05 |
ENST00000334166.4 |
LYPD6 |
LY6/PLAUR domain containing 6 |
| chr11_-_26743546 | 0.05 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_+_214161854 | 0.05 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr16_-_49698136 | 0.05 |
ENST00000535559.1 |
ZNF423 |
zinc finger protein 423 |
| chr2_+_152266392 | 0.05 |
ENST00000444746.2 ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr2_-_87248975 | 0.05 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr19_+_44037334 | 0.05 |
ENST00000314228.5 |
ZNF575 |
zinc finger protein 575 |
| chr4_-_186456652 | 0.05 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr17_+_4613918 | 0.05 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chr2_+_103089756 | 0.05 |
ENST00000295269.4 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr7_+_106505696 | 0.05 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr6_-_165989936 | 0.05 |
ENST00000354448.4 |
PDE10A |
phosphodiesterase 10A |
| chr1_-_120354079 | 0.05 |
ENST00000354219.1 ENST00000369401.4 ENST00000256585.5 |
REG4 |
regenerating islet-derived family, member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 1.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 1.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0030300 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.1 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 1.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0000406 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |