Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for FOXA3_FOXC2
Z-value: 0.17
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
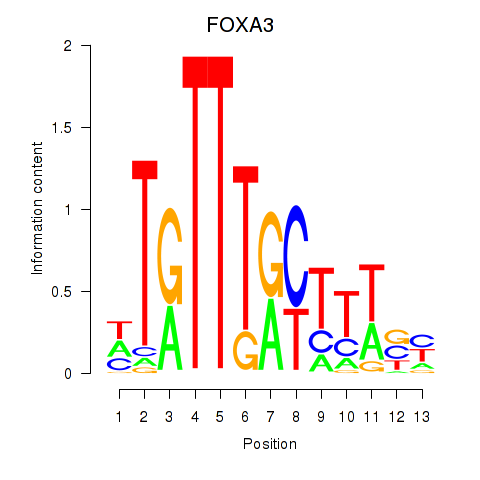
FOXA3
|
ENSG00000170608.2 | FOXA3 |
|
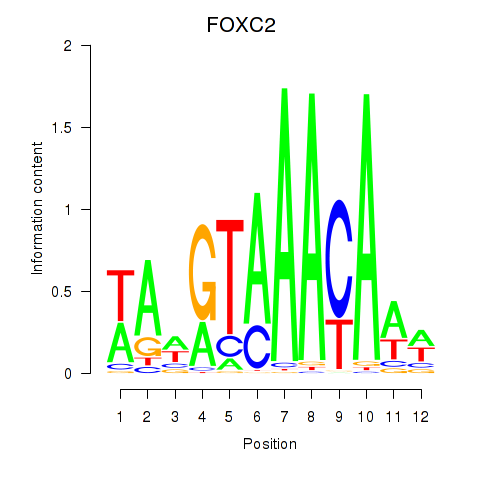
FOXC2
|
ENSG00000176692.4 | FOXC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | 0.52 | 1.9e-01 | Click! |
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.27 | 5.1e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_128175997 | 0.30 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr8_+_97597148 | 0.25 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr17_-_66951474 | 0.25 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr5_+_156712372 | 0.24 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_25616519 | 0.23 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr12_-_15038779 | 0.23 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chrX_+_85969626 | 0.23 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr9_-_95186739 | 0.22 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr17_-_19290483 | 0.20 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr5_+_40841276 | 0.17 |
ENST00000254691.5 |
CARD6 |
caspase recruitment domain family, member 6 |
| chr3_+_148447887 | 0.17 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr8_-_120651020 | 0.16 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr1_+_159141397 | 0.16 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr4_-_138453606 | 0.15 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr3_-_105588231 | 0.14 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr8_+_104384616 | 0.14 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr6_+_72922590 | 0.14 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr6_+_72922505 | 0.14 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr17_-_29641084 | 0.13 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr12_+_59989918 | 0.13 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_92539614 | 0.13 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr6_-_88875654 | 0.12 |
ENST00000535130.1 |
CNR1 |
cannabinoid receptor 1 (brain) |
| chr10_+_95848824 | 0.12 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr3_+_69985734 | 0.12 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_175711133 | 0.12 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr3_+_114012819 | 0.11 |
ENST00000383671.3 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
| chr2_+_88047606 | 0.11 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr14_+_21156915 | 0.11 |
ENST00000397990.4 ENST00000555597.1 |
ANG RNASE4 |
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr20_+_31823792 | 0.10 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr10_+_70847852 | 0.10 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr2_+_179318295 | 0.10 |
ENST00000442710.1 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr4_-_186696425 | 0.10 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr7_+_106809406 | 0.09 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr4_-_138453559 | 0.09 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr16_-_87970122 | 0.09 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr10_-_49482907 | 0.09 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr6_+_101846664 | 0.08 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr11_-_102401469 | 0.08 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr7_+_138943265 | 0.08 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr3_-_186080012 | 0.08 |
ENST00000544847.1 ENST00000265022.3 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
| chr17_-_26694979 | 0.08 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr3_+_69985792 | 0.07 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr17_-_26695013 | 0.07 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr12_+_59989791 | 0.07 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr18_-_52626622 | 0.07 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr6_+_122793058 | 0.07 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr7_+_134576317 | 0.06 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr6_-_117747015 | 0.06 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr1_+_104293028 | 0.06 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr3_-_114343039 | 0.06 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_-_3350614 | 0.06 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr16_-_67597789 | 0.06 |
ENST00000605277.1 |
CTD-2012K14.6 |
CTD-2012K14.6 |
| chr2_-_145275228 | 0.06 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_-_120354079 | 0.06 |
ENST00000354219.1 ENST00000369401.4 ENST00000256585.5 |
REG4 |
regenerating islet-derived family, member 4 |
| chr7_+_151038850 | 0.05 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr2_+_109223595 | 0.05 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr5_+_140710061 | 0.05 |
ENST00000517417.1 ENST00000378105.3 |
PCDHGA1 |
protocadherin gamma subfamily A, 1 |
| chr2_+_220042933 | 0.05 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chrX_+_9431324 | 0.05 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr7_+_134430212 | 0.05 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr14_-_101036119 | 0.05 |
ENST00000355173.2 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr5_+_101569696 | 0.05 |
ENST00000597120.1 |
AC008948.1 |
AC008948.1 |
| chr10_-_118032697 | 0.05 |
ENST00000439649.3 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr4_+_88754113 | 0.05 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr3_-_187455680 | 0.05 |
ENST00000438077.1 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr4_+_88754069 | 0.05 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr20_+_45338126 | 0.05 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr1_-_57431679 | 0.04 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr3_+_119316689 | 0.04 |
ENST00000273371.4 |
PLA1A |
phospholipase A1 member A |
| chr3_+_119316721 | 0.04 |
ENST00000488919.1 ENST00000495992.1 |
PLA1A |
phospholipase A1 member A |
| chr4_+_86396265 | 0.04 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_+_175260514 | 0.04 |
ENST00000424069.1 ENST00000427038.1 |
SCRN3 |
secernin 3 |
| chr1_+_104159999 | 0.04 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr1_+_101361782 | 0.04 |
ENST00000357650.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr6_+_108882069 | 0.04 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr7_+_142829162 | 0.04 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr4_-_70826725 | 0.04 |
ENST00000353151.3 |
CSN2 |
casein beta |
| chr2_-_32490859 | 0.04 |
ENST00000404025.2 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr2_+_135596106 | 0.04 |
ENST00000356140.5 |
ACMSD |
aminocarboxymuconate semialdehyde decarboxylase |
| chr1_-_207095212 | 0.04 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr1_+_18957500 | 0.04 |
ENST00000375375.3 |
PAX7 |
paired box 7 |
| chr2_+_175260451 | 0.04 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr11_-_82708519 | 0.04 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr1_-_177939348 | 0.04 |
ENST00000464631.2 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
| chr4_-_21950356 | 0.04 |
ENST00000447367.2 ENST00000382152.2 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr2_+_135596180 | 0.04 |
ENST00000283054.4 ENST00000392928.1 |
ACMSD |
aminocarboxymuconate semialdehyde decarboxylase |
| chr6_+_12717892 | 0.04 |
ENST00000379350.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr1_-_169555779 | 0.04 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr14_-_74551096 | 0.03 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr17_+_56232494 | 0.03 |
ENST00000268912.5 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
| chr5_+_140529630 | 0.03 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr20_-_43150601 | 0.03 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr2_-_106054952 | 0.03 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr3_-_71294304 | 0.03 |
ENST00000498215.1 |
FOXP1 |
forkhead box P1 |
| chr14_+_74034310 | 0.03 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr8_+_75736761 | 0.03 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr11_-_82708435 | 0.03 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr20_+_31870927 | 0.03 |
ENST00000253354.1 |
BPIFB1 |
BPI fold containing family B, member 1 |
| chr3_+_50712672 | 0.03 |
ENST00000266037.9 |
DOCK3 |
dedicator of cytokinesis 3 |
| chr2_+_166150541 | 0.03 |
ENST00000283256.6 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_+_21666633 | 0.03 |
ENST00000606851.1 |
CASC15 |
cancer susceptibility candidate 15 (non-protein coding) |
| chr8_+_52730143 | 0.03 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr6_-_87804815 | 0.03 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chrX_+_65382433 | 0.03 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr8_+_95908041 | 0.03 |
ENST00000396113.1 ENST00000519136.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr16_-_15474904 | 0.03 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr18_+_61575200 | 0.03 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr12_-_71551652 | 0.03 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr14_-_74551172 | 0.03 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr2_+_169659121 | 0.03 |
ENST00000397206.2 ENST00000397209.2 ENST00000421711.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chr17_-_39471947 | 0.03 |
ENST00000334202.3 |
KRTAP17-1 |
keratin associated protein 17-1 |
| chr5_-_159846399 | 0.03 |
ENST00000297151.4 |
SLU7 |
SLU7 splicing factor homolog (S. cerevisiae) |
| chr21_+_17791648 | 0.03 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr16_+_14802801 | 0.03 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr2_+_169658928 | 0.03 |
ENST00000317647.7 ENST00000445023.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chr6_-_122792919 | 0.02 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr12_+_58087901 | 0.02 |
ENST00000315970.7 ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9 |
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr8_+_67579807 | 0.02 |
ENST00000519289.1 ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3 C8orf44 |
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr14_-_94789663 | 0.02 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr15_+_60296421 | 0.02 |
ENST00000396057.4 |
FOXB1 |
forkhead box B1 |
| chr11_+_8040739 | 0.02 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr2_-_183291741 | 0.02 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_3449330 | 0.02 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_-_25479811 | 0.02 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr1_+_246729724 | 0.02 |
ENST00000366513.4 ENST00000366512.3 |
CNST |
consortin, connexin sorting protein |
| chr14_-_24664540 | 0.02 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr17_-_46688334 | 0.02 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr20_+_44519948 | 0.02 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chr20_+_9494987 | 0.02 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr7_+_134464376 | 0.02 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr14_-_70263979 | 0.02 |
ENST00000216540.4 |
SLC10A1 |
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr3_+_141106643 | 0.02 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr9_-_70490107 | 0.02 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr13_-_39564993 | 0.02 |
ENST00000423210.1 |
STOML3 |
stomatin (EPB72)-like 3 |
| chr19_+_50380682 | 0.02 |
ENST00000221543.5 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr1_+_40974431 | 0.02 |
ENST00000296380.4 ENST00000432259.1 ENST00000418186.1 |
EXO5 |
exonuclease 5 |
| chr7_+_106505696 | 0.02 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_146696901 | 0.02 |
ENST00000369272.3 ENST00000441068.2 |
FMO5 |
flavin containing monooxygenase 5 |
| chr12_-_92536433 | 0.02 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr2_-_182545603 | 0.02 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr2_-_65593784 | 0.02 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr7_-_107443652 | 0.02 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr19_+_50380917 | 0.02 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr1_+_18958008 | 0.02 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr1_+_114471809 | 0.02 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr2_+_68592305 | 0.02 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr21_+_17791838 | 0.02 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr3_-_168864427 | 0.02 |
ENST00000468789.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr1_+_207277590 | 0.02 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr9_+_70856397 | 0.02 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr7_+_116654935 | 0.02 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr20_+_44520009 | 0.02 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr4_+_148653206 | 0.02 |
ENST00000336498.3 |
ARHGAP10 |
Rho GTPase activating protein 10 |
| chr21_-_43786634 | 0.02 |
ENST00000291527.2 |
TFF1 |
trefoil factor 1 |
| chr7_+_134464414 | 0.02 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr2_-_87248975 | 0.02 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chrX_+_10124977 | 0.02 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr15_-_40600026 | 0.02 |
ENST00000456256.2 ENST00000557821.1 |
PLCB2 |
phospholipase C, beta 2 |
| chr14_+_52164820 | 0.02 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr1_+_12806141 | 0.02 |
ENST00000288048.5 |
C1orf158 |
chromosome 1 open reading frame 158 |
| chr3_+_130569592 | 0.02 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_-_18902106 | 0.02 |
ENST00000542601.2 ENST00000425807.1 ENST00000222271.2 |
COMP |
cartilage oligomeric matrix protein |
| chr12_+_72080253 | 0.02 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chrX_+_90689810 | 0.02 |
ENST00000312600.3 |
PABPC5 |
poly(A) binding protein, cytoplasmic 5 |
| chr10_+_35484053 | 0.02 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr10_-_21435488 | 0.02 |
ENST00000534331.1 ENST00000529198.1 ENST00000377118.4 |
C10orf113 |
chromosome 10 open reading frame 113 |
| chr10_-_31288398 | 0.02 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr2_-_175260368 | 0.02 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chr2_-_145275211 | 0.02 |
ENST00000462355.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr7_-_81399411 | 0.02 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_4285923 | 0.02 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr2_-_188312971 | 0.02 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr3_-_116163830 | 0.02 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr8_+_95907993 | 0.02 |
ENST00000523378.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr18_-_31803435 | 0.02 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr14_-_23526739 | 0.02 |
ENST00000397359.3 ENST00000487137.2 |
CDH24 |
cadherin 24, type 2 |
| chr1_+_84609944 | 0.02 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_-_112575687 | 0.02 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr7_+_106505912 | 0.01 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr3_+_159570722 | 0.01 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr6_+_30103885 | 0.01 |
ENST00000396581.1 |
TRIM40 |
tripartite motif containing 40 |
| chr6_-_26056695 | 0.01 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr1_-_43855479 | 0.01 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr1_+_151739131 | 0.01 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr15_+_71185148 | 0.01 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr8_+_55528627 | 0.01 |
ENST00000220676.1 |
RP1 |
retinitis pigmentosa 1 (autosomal dominant) |
| chr19_+_782755 | 0.01 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr18_-_53070913 | 0.01 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr14_+_23654525 | 0.01 |
ENST00000399910.1 ENST00000492621.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr18_+_21033239 | 0.01 |
ENST00000581585.1 ENST00000577501.1 |
RIOK3 |
RIO kinase 3 |
| chrX_-_80457385 | 0.01 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr1_+_43855560 | 0.01 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr4_-_170679024 | 0.01 |
ENST00000393381.2 |
C4orf27 |
chromosome 4 open reading frame 27 |
| chr4_+_86396321 | 0.01 |
ENST00000503995.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr17_-_39203519 | 0.01 |
ENST00000542137.1 ENST00000391419.3 |
KRTAP2-1 |
keratin associated protein 2-1 |
| chr15_-_50411412 | 0.01 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr12_+_93096619 | 0.01 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr1_-_150602035 | 0.01 |
ENST00000503241.1 ENST00000369016.4 ENST00000339643.5 ENST00000271690.8 ENST00000356527.5 ENST00000362052.7 ENST00000503345.1 ENST00000369014.5 ENST00000369009.3 |
ENSA |
endosulfine alpha |
| chr1_+_61548225 | 0.01 |
ENST00000371187.3 |
NFIA |
nuclear factor I/A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.0 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |