Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for FOXC1
Z-value: 0.09
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | FOXC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | 0.55 | 1.6e-01 | Click! |
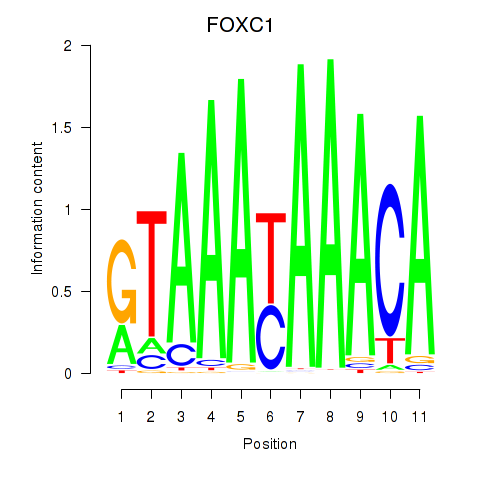
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_19290483 | 1.07 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr8_-_93107443 | 0.97 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_-_52626622 | 0.63 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr8_+_75736761 | 0.57 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr13_-_86373536 | 0.56 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr8_-_108510224 | 0.51 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr2_+_166095898 | 0.50 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr8_-_93107827 | 0.43 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_-_93107696 | 0.43 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_+_101846664 | 0.42 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chrX_+_100743031 | 0.41 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chr4_-_186696425 | 0.36 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_+_84563295 | 0.34 |
ENST00000369687.1 |
RIPPLY2 |
ripply transcriptional repressor 2 |
| chr1_+_155099927 | 0.33 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr9_-_89562104 | 0.30 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr1_+_197886461 | 0.30 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr3_-_114343039 | 0.29 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_+_97597148 | 0.28 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr11_-_108464465 | 0.26 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr4_+_86396265 | 0.26 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr8_+_104384616 | 0.24 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr8_-_19540086 | 0.24 |
ENST00000332246.6 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_+_31496809 | 0.24 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr2_-_161056802 | 0.24 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056762 | 0.22 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr10_-_4285923 | 0.22 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr3_-_57326704 | 0.21 |
ENST00000487349.1 ENST00000389601.3 |
ASB14 |
ankyrin repeat and SOCS box containing 14 |
| chr11_-_108464321 | 0.21 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr5_+_40841276 | 0.21 |
ENST00000254691.5 |
CARD6 |
caspase recruitment domain family, member 6 |
| chr18_+_19749386 | 0.20 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr18_-_53303123 | 0.20 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr1_+_18958008 | 0.19 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr3_-_114477962 | 0.18 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_-_89744457 | 0.18 |
ENST00000395002.2 |
FAM13A |
family with sequence similarity 13, member A |
| chr8_-_80993010 | 0.18 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr8_-_134309823 | 0.18 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr3_-_114477787 | 0.18 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr21_+_17791648 | 0.18 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr1_+_220701456 | 0.18 |
ENST00000366918.4 ENST00000402574.1 |
MARK1 |
MAP/microtubule affinity-regulating kinase 1 |
| chr21_+_17791838 | 0.17 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr8_-_134309335 | 0.17 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr4_+_86396321 | 0.15 |
ENST00000503995.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr5_-_39425290 | 0.15 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425222 | 0.15 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_+_114709999 | 0.14 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_118032697 | 0.14 |
ENST00000439649.3 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr1_-_228135599 | 0.14 |
ENST00000272164.5 |
WNT9A |
wingless-type MMTV integration site family, member 9A |
| chr19_-_51141196 | 0.14 |
ENST00000338916.4 |
SYT3 |
synaptotagmin III |
| chr19_-_36523709 | 0.14 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr3_-_15382875 | 0.13 |
ENST00000408919.3 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr7_+_129906660 | 0.13 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chrX_-_77914825 | 0.13 |
ENST00000321110.1 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
| chr6_+_10528560 | 0.13 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr3_+_114012819 | 0.13 |
ENST00000383671.3 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
| chr3_-_139195350 | 0.13 |
ENST00000232217.2 |
RBP2 |
retinol binding protein 2, cellular |
| chr10_+_114710211 | 0.12 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr21_+_17792672 | 0.12 |
ENST00000602620.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr5_-_39425068 | 0.12 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_123241908 | 0.12 |
ENST00000434204.1 ENST00000437535.1 ENST00000451215.1 |
ASB15 |
ankyrin repeat and SOCS box containing 15 |
| chr2_+_128175997 | 0.12 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr12_+_43086018 | 0.12 |
ENST00000550177.1 |
RP11-25I15.3 |
RP11-25I15.3 |
| chr10_-_61122220 | 0.12 |
ENST00000422313.2 ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C |
family with sequence similarity 13, member C |
| chr12_-_10251539 | 0.11 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr15_+_67418047 | 0.11 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr10_+_52751010 | 0.11 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr18_-_53070913 | 0.11 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr16_+_72459838 | 0.11 |
ENST00000564508.1 |
AC004158.3 |
AC004158.3 |
| chr2_+_210517895 | 0.11 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr7_+_80231466 | 0.11 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr5_-_146258205 | 0.11 |
ENST00000394413.3 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_153931052 | 0.10 |
ENST00000368630.3 ENST00000368633.1 |
CRTC2 |
CREB regulated transcription coactivator 2 |
| chrX_+_22050546 | 0.10 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr8_-_23261589 | 0.09 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr8_-_72274095 | 0.09 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr19_-_40919271 | 0.09 |
ENST00000291825.7 ENST00000324001.7 |
PRX |
periaxin |
| chr22_-_38699003 | 0.09 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chr8_-_72274467 | 0.09 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr17_+_7533439 | 0.09 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr17_+_8924837 | 0.09 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr2_+_171571827 | 0.08 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr14_-_92572894 | 0.08 |
ENST00000532032.1 ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3 |
ataxin 3 |
| chr5_-_36301984 | 0.08 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr21_-_34186006 | 0.08 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr15_-_30114622 | 0.08 |
ENST00000495972.2 ENST00000346128.6 |
TJP1 |
tight junction protein 1 |
| chr11_+_8040739 | 0.07 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr12_+_131438443 | 0.07 |
ENST00000261654.5 |
GPR133 |
G protein-coupled receptor 133 |
| chr6_+_47666275 | 0.07 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr4_-_103266626 | 0.07 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr19_+_36024310 | 0.07 |
ENST00000222286.4 |
GAPDHS |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr7_+_79998864 | 0.07 |
ENST00000435819.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr17_-_39165366 | 0.07 |
ENST00000391588.1 |
KRTAP3-1 |
keratin associated protein 3-1 |
| chr10_+_112631547 | 0.07 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr2_-_32490859 | 0.07 |
ENST00000404025.2 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr11_+_10476851 | 0.07 |
ENST00000396553.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr18_+_21032781 | 0.07 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr22_-_46659219 | 0.07 |
ENST00000253255.5 |
PKDREJ |
polycystin (PKD) family receptor for egg jelly |
| chr19_-_52255107 | 0.07 |
ENST00000595042.1 ENST00000304748.4 |
FPR1 |
formyl peptide receptor 1 |
| chr12_+_10365404 | 0.07 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr2_-_72375167 | 0.06 |
ENST00000001146.2 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr14_+_23340822 | 0.06 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr4_-_105416039 | 0.06 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr7_-_139876812 | 0.06 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr19_-_47734448 | 0.06 |
ENST00000439096.2 |
BBC3 |
BCL2 binding component 3 |
| chr3_+_189349162 | 0.05 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr18_+_21033239 | 0.05 |
ENST00000581585.1 ENST00000577501.1 |
RIOK3 |
RIO kinase 3 |
| chr11_-_62911693 | 0.05 |
ENST00000417740.1 ENST00000326192.5 |
SLC22A24 |
solute carrier family 22, member 24 |
| chr2_-_27886676 | 0.05 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr12_+_55413721 | 0.05 |
ENST00000242994.3 |
NEUROD4 |
neuronal differentiation 4 |
| chr17_-_37764128 | 0.05 |
ENST00000302584.4 |
NEUROD2 |
neuronal differentiation 2 |
| chr15_-_72563585 | 0.05 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr2_-_214013353 | 0.05 |
ENST00000451136.2 ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr8_-_141931287 | 0.05 |
ENST00000517887.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr8_+_52730143 | 0.05 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chrX_+_38211777 | 0.05 |
ENST00000039007.4 |
OTC |
ornithine carbamoyltransferase |
| chr16_+_6069586 | 0.05 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_+_46638805 | 0.05 |
ENST00000434074.1 ENST00000312040.4 ENST00000451945.1 |
ATG13 |
autophagy related 13 |
| chrX_-_117107680 | 0.05 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr11_+_46639071 | 0.05 |
ENST00000580238.1 ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13 |
autophagy related 13 |
| chr19_-_36523529 | 0.05 |
ENST00000593074.1 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr12_+_99038919 | 0.05 |
ENST00000551964.1 |
APAF1 |
apoptotic peptidase activating factor 1 |
| chr2_-_8977714 | 0.05 |
ENST00000319688.5 ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220 |
kinase D-interacting substrate, 220kDa |
| chr13_-_36050819 | 0.04 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr6_-_10747802 | 0.04 |
ENST00000606522.1 ENST00000606652.1 |
RP11-421M1.8 |
RP11-421M1.8 |
| chr1_-_246729544 | 0.04 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr1_+_18957500 | 0.04 |
ENST00000375375.3 |
PAX7 |
paired box 7 |
| chr3_-_48481434 | 0.04 |
ENST00000395694.2 ENST00000447018.1 ENST00000442740.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr17_-_27503770 | 0.04 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr3_-_48481518 | 0.04 |
ENST00000412398.2 ENST00000395696.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr2_+_162272605 | 0.04 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr12_+_122516626 | 0.04 |
ENST00000319080.7 |
MLXIP |
MLX interacting protein |
| chr2_+_207024306 | 0.04 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr16_+_56969284 | 0.04 |
ENST00000568358.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr14_-_61190754 | 0.04 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr6_-_32083106 | 0.04 |
ENST00000442721.1 |
TNXB |
tenascin XB |
| chr11_+_112832090 | 0.04 |
ENST00000533760.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr3_-_141868357 | 0.04 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_128902729 | 0.04 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr2_-_27886460 | 0.04 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr12_-_10251603 | 0.04 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr2_+_113885138 | 0.04 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr7_-_86849025 | 0.04 |
ENST00000257637.3 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr3_-_16524357 | 0.03 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chrX_-_117107542 | 0.03 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr3_-_128902759 | 0.03 |
ENST00000422453.2 ENST00000504813.1 ENST00000512338.1 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr12_+_93096759 | 0.03 |
ENST00000544406.2 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr1_+_246729724 | 0.03 |
ENST00000366513.4 ENST00000366512.3 |
CNST |
consortin, connexin sorting protein |
| chr17_+_57642886 | 0.03 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr15_+_58430368 | 0.03 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr15_+_58430567 | 0.03 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr10_+_71561649 | 0.03 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr17_+_57408994 | 0.03 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr15_+_80351977 | 0.03 |
ENST00000559157.1 ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr2_+_44502597 | 0.03 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr12_+_93096619 | 0.03 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr3_-_11623804 | 0.03 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr8_-_114449112 | 0.03 |
ENST00000455883.2 ENST00000352409.3 ENST00000297405.5 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr6_-_5260963 | 0.03 |
ENST00000464010.1 ENST00000468929.1 ENST00000480566.1 |
LYRM4 |
LYR motif containing 4 |
| chr1_+_25664408 | 0.03 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr5_+_161494521 | 0.03 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_+_181845843 | 0.03 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr19_-_4831701 | 0.03 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr2_+_28974603 | 0.03 |
ENST00000441461.1 ENST00000358506.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr16_+_6069072 | 0.03 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr10_+_95517616 | 0.03 |
ENST00000371418.4 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr2_+_28974668 | 0.03 |
ENST00000296122.6 ENST00000395366.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr8_-_145754428 | 0.02 |
ENST00000527462.1 ENST00000313465.5 ENST00000524821.1 |
C8orf82 |
chromosome 8 open reading frame 82 |
| chr11_+_34654011 | 0.02 |
ENST00000531794.1 |
EHF |
ets homologous factor |
| chr21_+_38071430 | 0.02 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr5_-_146302078 | 0.02 |
ENST00000508545.2 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr10_+_95517566 | 0.02 |
ENST00000542308.1 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr1_+_73771844 | 0.02 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr17_-_48546232 | 0.02 |
ENST00000258969.4 |
CHAD |
chondroadherin |
| chr16_+_8715536 | 0.02 |
ENST00000563958.1 ENST00000381920.3 ENST00000564554.1 |
METTL22 |
methyltransferase like 22 |
| chr16_+_8715574 | 0.02 |
ENST00000561758.1 |
METTL22 |
methyltransferase like 22 |
| chr6_-_39399087 | 0.02 |
ENST00000229913.5 ENST00000541946.1 ENST00000394362.1 |
KIF6 |
kinesin family member 6 |
| chrX_+_85969626 | 0.02 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr15_-_55657428 | 0.02 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr5_+_159614374 | 0.02 |
ENST00000393980.4 |
FABP6 |
fatty acid binding protein 6, ileal |
| chr14_-_20774092 | 0.02 |
ENST00000423949.2 ENST00000553828.1 ENST00000258821.3 |
TTC5 |
tetratricopeptide repeat domain 5 |
| chr6_+_119215308 | 0.02 |
ENST00000229595.5 |
ASF1A |
anti-silencing function 1A histone chaperone |
| chr6_+_10747986 | 0.02 |
ENST00000379542.5 |
TMEM14B |
transmembrane protein 14B |
| chr4_-_74486347 | 0.02 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_+_42120283 | 0.02 |
ENST00000542534.2 ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B JMJD7 JMJD7-PLA2G4B |
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr11_-_66112555 | 0.02 |
ENST00000425825.2 ENST00000359957.3 |
BRMS1 |
breast cancer metastasis suppressor 1 |
| chr1_-_149982624 | 0.01 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
| chr4_-_74486109 | 0.01 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_60933634 | 0.01 |
ENST00000505642.1 |
C5orf64 |
chromosome 5 open reading frame 64 |
| chr7_-_14028488 | 0.01 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr3_-_57233966 | 0.01 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr3_-_157221128 | 0.01 |
ENST00000392833.2 ENST00000362010.2 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chrY_+_59100480 | 0.01 |
ENSTR0000302805.2 |
SPRY3 |
sprouty homolog 3 (Drosophila) |
| chr7_+_151038850 | 0.01 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr15_-_50411412 | 0.01 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr15_+_80351910 | 0.01 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr14_+_37126765 | 0.01 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr1_+_174846570 | 0.01 |
ENST00000392064.2 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr10_+_71561630 | 0.01 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr13_+_27825706 | 0.01 |
ENST00000272274.4 ENST00000319826.4 ENST00000326092.4 |
RPL21 |
ribosomal protein L21 |
| chr4_-_147443043 | 0.01 |
ENST00000394059.4 ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7 |
solute carrier family 10, member 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.2 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 1.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:0036446 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0010482 | ectoderm and mesoderm interaction(GO:0007499) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |