Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
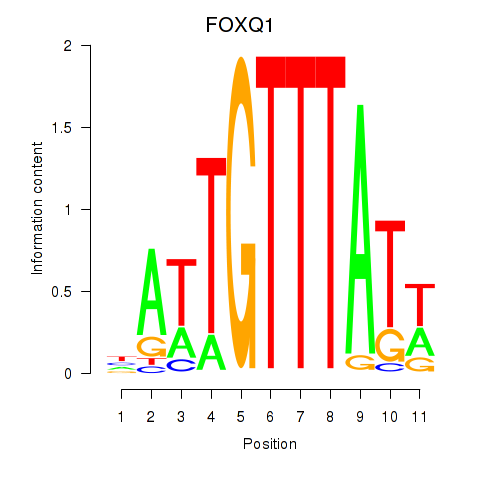
Results for FOXQ1
Z-value: 0.81
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.4 | FOXQ1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg19_v2_chr6_+_1312675_1312701 | -0.56 | 1.5e-01 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 1.90 |
ENST00000547937.1 |
DCN |
decorin |
| chr3_+_157154578 | 1.66 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr4_-_70626314 | 1.18 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr5_+_140529630 | 1.04 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr4_-_70626430 | 0.99 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr2_+_152214098 | 0.96 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_188419200 | 0.94 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_-_72566613 | 0.94 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr2_-_188419078 | 0.91 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_-_20575959 | 0.83 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr2_+_217082311 | 0.76 |
ENST00000597904.1 |
RP11-566E18.3 |
RP11-566E18.3 |
| chr3_+_69985734 | 0.57 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr12_+_1738363 | 0.57 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr10_+_70847852 | 0.54 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr6_-_87804815 | 0.52 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr5_-_111091948 | 0.52 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr8_+_70404996 | 0.51 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr5_-_41510725 | 0.48 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr10_-_79397202 | 0.46 |
ENST00000372437.1 ENST00000372408.2 ENST00000372403.4 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr18_-_70532906 | 0.43 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_+_145524891 | 0.42 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr21_+_25801041 | 0.42 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr2_+_201450591 | 0.39 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr8_+_79428539 | 0.39 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr5_-_41510656 | 0.39 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_+_10292308 | 0.38 |
ENST00000377081.1 |
KIF1B |
kinesin family member 1B |
| chr20_+_56964169 | 0.37 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_140743859 | 0.37 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr1_-_24194771 | 0.37 |
ENST00000374479.3 |
FUCA1 |
fucosidase, alpha-L- 1, tissue |
| chr20_+_34802295 | 0.35 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr9_-_73477826 | 0.35 |
ENST00000396285.1 ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr6_-_169364429 | 0.35 |
ENST00000444586.1 |
RP3-495K2.3 |
RP3-495K2.3 |
| chr1_-_27998689 | 0.34 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr10_-_49812997 | 0.32 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr5_+_140571902 | 0.32 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr10_-_49813090 | 0.32 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_104159999 | 0.32 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr1_+_39876151 | 0.31 |
ENST00000530275.1 |
KIAA0754 |
KIAA0754 |
| chr2_-_85555385 | 0.31 |
ENST00000377386.3 |
TGOLN2 |
trans-golgi network protein 2 |
| chr11_-_27723158 | 0.31 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr14_+_50999744 | 0.30 |
ENST00000441560.2 |
ATL1 |
atlastin GTPase 1 |
| chr18_+_7754957 | 0.30 |
ENST00000400053.4 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr20_-_3185279 | 0.30 |
ENST00000354488.3 ENST00000380201.2 |
DDRGK1 |
DDRGK domain containing 1 |
| chr20_+_12989596 | 0.29 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr5_+_140625147 | 0.29 |
ENST00000231173.3 |
PCDHB15 |
protocadherin beta 15 |
| chr16_+_22517166 | 0.29 |
ENST00000356156.3 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr6_-_88875654 | 0.28 |
ENST00000535130.1 |
CNR1 |
cannabinoid receptor 1 (brain) |
| chr2_+_108994466 | 0.28 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr5_-_159846399 | 0.27 |
ENST00000297151.4 |
SLU7 |
SLU7 splicing factor homolog (S. cerevisiae) |
| chr14_+_51026743 | 0.27 |
ENST00000358385.6 ENST00000357032.3 ENST00000354525.4 |
ATL1 |
atlastin GTPase 1 |
| chr5_-_24645078 | 0.27 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr2_-_192711968 | 0.27 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr12_-_122879969 | 0.27 |
ENST00000540304.1 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
| chr1_+_240177627 | 0.27 |
ENST00000447095.1 |
FMN2 |
formin 2 |
| chr1_-_227505289 | 0.26 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_+_10124977 | 0.26 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr5_-_88119580 | 0.26 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr4_-_119274121 | 0.25 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr3_+_82035289 | 0.25 |
ENST00000470263.1 ENST00000494340.1 |
RP11-260O18.1 |
RP11-260O18.1 |
| chr12_-_71551652 | 0.25 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr20_+_30102231 | 0.25 |
ENST00000335574.5 ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13 |
histocompatibility (minor) 13 |
| chr17_-_19648683 | 0.25 |
ENST00000573368.1 ENST00000457500.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_178976996 | 0.24 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr2_-_203735976 | 0.24 |
ENST00000435143.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr2_+_108994633 | 0.24 |
ENST00000409309.3 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_+_197170592 | 0.24 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr3_+_69985792 | 0.24 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr7_-_50860565 | 0.24 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr11_-_124670550 | 0.23 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr9_-_123239632 | 0.23 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr5_-_139937895 | 0.23 |
ENST00000336283.6 |
SRA1 |
steroid receptor RNA activator 1 |
| chr7_+_134528635 | 0.23 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr14_+_88471468 | 0.23 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr1_+_162602244 | 0.23 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr1_-_54405773 | 0.23 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr1_-_145826450 | 0.22 |
ENST00000462900.2 |
GPR89A |
G protein-coupled receptor 89A |
| chr7_+_142985308 | 0.22 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr4_+_165675269 | 0.22 |
ENST00000507311.1 |
RP11-294O2.2 |
RP11-294O2.2 |
| chr3_+_577893 | 0.22 |
ENST00000420823.1 ENST00000442809.1 |
AC090044.1 |
AC090044.1 |
| chr6_+_116575329 | 0.21 |
ENST00000430252.2 ENST00000540275.1 ENST00000448740.2 |
DSE RP3-486I3.7 |
dermatan sulfate epimerase RP3-486I3.7 |
| chr1_+_174844645 | 0.21 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr5_+_72143988 | 0.21 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr3_+_152017924 | 0.21 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr1_+_145883868 | 0.21 |
ENST00000447947.2 |
GPR89C |
G protein-coupled receptor 89C |
| chr15_-_39486510 | 0.20 |
ENST00000560743.1 |
RP11-265N7.1 |
RP11-265N7.1 |
| chr5_-_13944652 | 0.20 |
ENST00000265104.4 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
| chr4_-_111119804 | 0.20 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr12_+_122880045 | 0.20 |
ENST00000539034.1 ENST00000535976.1 |
RP11-450K4.1 |
RP11-450K4.1 |
| chr15_+_78730622 | 0.20 |
ENST00000560440.1 |
IREB2 |
iron-responsive element binding protein 2 |
| chr2_+_109237717 | 0.20 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr5_+_140718396 | 0.20 |
ENST00000394576.2 |
PCDHGA2 |
protocadherin gamma subfamily A, 2 |
| chr15_+_41549105 | 0.20 |
ENST00000560965.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr10_+_89420706 | 0.19 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr12_-_92539614 | 0.19 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr14_-_74417096 | 0.19 |
ENST00000286544.3 |
FAM161B |
family with sequence similarity 161, member B |
| chr11_+_20044096 | 0.19 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr10_+_15085895 | 0.18 |
ENST00000378228.3 |
OLAH |
oleoyl-ACP hydrolase |
| chr2_+_197577841 | 0.18 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr1_+_114471809 | 0.18 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr7_+_48211048 | 0.18 |
ENST00000435803.1 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr2_+_158114051 | 0.18 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr11_+_6947647 | 0.17 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr2_+_179317994 | 0.17 |
ENST00000375129.4 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr9_+_2029019 | 0.17 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_63953415 | 0.17 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr9_-_123812542 | 0.17 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr10_+_15085936 | 0.17 |
ENST00000378217.3 |
OLAH |
oleoyl-ACP hydrolase |
| chrM_+_5824 | 0.17 |
ENST00000361624.2 |
MT-CO1 |
mitochondrially encoded cytochrome c oxidase I |
| chr3_-_12587055 | 0.17 |
ENST00000564146.3 |
C3orf83 |
chromosome 3 open reading frame 83 |
| chr20_+_48429233 | 0.17 |
ENST00000417961.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chrX_-_107975917 | 0.17 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr1_-_182921119 | 0.17 |
ENST00000423786.1 |
SHCBP1L |
SHC SH2-domain binding protein 1-like |
| chr1_+_61542922 | 0.17 |
ENST00000407417.3 |
NFIA |
nuclear factor I/A |
| chr19_+_50433476 | 0.16 |
ENST00000596658.1 |
ATF5 |
activating transcription factor 5 |
| chr16_+_22501658 | 0.16 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr17_-_295730 | 0.16 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr20_+_48429356 | 0.16 |
ENST00000361573.2 ENST00000541138.1 ENST00000539601.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr19_-_58220517 | 0.16 |
ENST00000512439.2 ENST00000426889.1 |
ZNF154 |
zinc finger protein 154 |
| chr18_-_77276057 | 0.15 |
ENST00000597412.1 |
AC018445.1 |
Uncharacterized protein |
| chr20_-_43150601 | 0.15 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr6_-_31648150 | 0.15 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr2_-_106054952 | 0.15 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr17_-_33864772 | 0.15 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr7_-_72993033 | 0.15 |
ENST00000305632.5 |
TBL2 |
transducin (beta)-like 2 |
| chr4_-_23735183 | 0.15 |
ENST00000507666.1 |
RP11-380P13.2 |
RP11-380P13.2 |
| chrX_-_71525742 | 0.15 |
ENST00000450875.1 ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr14_-_73493784 | 0.15 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr5_+_140588269 | 0.15 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr5_+_102201722 | 0.15 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_74853897 | 0.15 |
ENST00000296028.3 |
PPBP |
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr19_+_782755 | 0.15 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr16_-_15474904 | 0.15 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr6_+_116691001 | 0.15 |
ENST00000537543.1 |
DSE |
dermatan sulfate epimerase |
| chr21_+_39628852 | 0.15 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr20_-_33732952 | 0.14 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr5_-_64920115 | 0.14 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr6_-_11382478 | 0.14 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr5_+_73109339 | 0.14 |
ENST00000296799.4 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr16_-_31076332 | 0.14 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr10_-_93669233 | 0.14 |
ENST00000311575.5 |
FGFBP3 |
fibroblast growth factor binding protein 3 |
| chr3_-_150920979 | 0.14 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr16_+_29991673 | 0.14 |
ENST00000416441.2 |
TAOK2 |
TAO kinase 2 |
| chr13_-_39564993 | 0.14 |
ENST00000423210.1 |
STOML3 |
stomatin (EPB72)-like 3 |
| chr20_+_30327063 | 0.14 |
ENST00000300403.6 ENST00000340513.4 |
TPX2 |
TPX2, microtubule-associated |
| chr1_+_206972215 | 0.14 |
ENST00000340758.2 |
IL19 |
interleukin 19 |
| chr19_-_57988871 | 0.14 |
ENST00000596831.1 ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9 ZNF772 |
Uncharacterized protein zinc finger protein 772 |
| chr10_-_31288398 | 0.13 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr3_-_151102529 | 0.13 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr11_+_73019282 | 0.13 |
ENST00000263674.3 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr12_-_49582978 | 0.13 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr11_-_26593677 | 0.13 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr6_-_114194483 | 0.13 |
ENST00000434296.2 |
RP1-249H1.4 |
RP1-249H1.4 |
| chr15_-_56757329 | 0.13 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr14_-_89878369 | 0.13 |
ENST00000553840.1 ENST00000556916.1 |
FOXN3 |
forkhead box N3 |
| chr3_-_119379719 | 0.13 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr11_-_62457371 | 0.13 |
ENST00000317449.4 |
LRRN4CL |
LRRN4 C-terminal like |
| chr15_+_91643442 | 0.13 |
ENST00000394232.1 |
SV2B |
synaptic vesicle glycoprotein 2B |
| chr20_+_36373032 | 0.13 |
ENST00000373473.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr11_-_47447970 | 0.13 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr12_-_47473425 | 0.13 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr2_-_203735586 | 0.12 |
ENST00000454326.1 ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr14_-_73493825 | 0.12 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr1_+_61330931 | 0.12 |
ENST00000371191.1 |
NFIA |
nuclear factor I/A |
| chr12_-_71551868 | 0.12 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr9_+_4662282 | 0.12 |
ENST00000381883.2 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr6_-_28973037 | 0.12 |
ENST00000377179.3 |
ZNF311 |
zinc finger protein 311 |
| chr12_+_109554386 | 0.12 |
ENST00000338432.7 |
ACACB |
acetyl-CoA carboxylase beta |
| chr2_+_132479948 | 0.12 |
ENST00000355171.4 |
C2orf27A |
chromosome 2 open reading frame 27A |
| chr3_+_107096188 | 0.12 |
ENST00000261058.1 |
CCDC54 |
coiled-coil domain containing 54 |
| chr16_+_2820912 | 0.12 |
ENST00000570539.1 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr16_-_90096309 | 0.12 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr1_+_114472222 | 0.12 |
ENST00000369558.1 ENST00000369561.4 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr12_-_47473557 | 0.12 |
ENST00000321382.3 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr4_-_82393009 | 0.11 |
ENST00000436139.2 |
RASGEF1B |
RasGEF domain family, member 1B |
| chr11_-_26593779 | 0.11 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr10_+_93683519 | 0.11 |
ENST00000265990.6 |
BTAF1 |
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chrX_-_50557302 | 0.11 |
ENST00000289292.7 |
SHROOM4 |
shroom family member 4 |
| chr13_+_52598827 | 0.11 |
ENST00000521776.2 |
UTP14C |
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chrX_+_88002226 | 0.11 |
ENST00000276127.4 ENST00000373111.1 |
CPXCR1 |
CPX chromosome region, candidate 1 |
| chr17_-_46690839 | 0.11 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chrX_-_54824673 | 0.11 |
ENST00000218436.6 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr5_+_64920543 | 0.11 |
ENST00000399438.3 ENST00000510585.2 |
TRAPPC13 CTC-534A2.2 |
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr4_-_82393052 | 0.11 |
ENST00000335927.7 ENST00000504863.1 ENST00000264400.2 |
RASGEF1B |
RasGEF domain family, member 1B |
| chr6_-_31681839 | 0.11 |
ENST00000409239.1 ENST00000461287.1 |
LY6G6E XXbac-BPG32J3.20 |
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr17_-_38574169 | 0.11 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr21_+_33671160 | 0.11 |
ENST00000303645.5 |
MRAP |
melanocortin 2 receptor accessory protein |
| chr20_-_34638841 | 0.11 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chr14_+_93389425 | 0.11 |
ENST00000216492.5 ENST00000334654.4 |
CHGA |
chromogranin A (parathyroid secretory protein 1) |
| chr1_+_236686717 | 0.11 |
ENST00000341872.6 ENST00000450372.2 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr17_+_30594823 | 0.10 |
ENST00000536287.1 |
RHBDL3 |
rhomboid, veinlet-like 3 (Drosophila) |
| chr15_-_50411412 | 0.10 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr1_+_214161854 | 0.10 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr1_-_182360498 | 0.10 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr5_+_118812294 | 0.10 |
ENST00000509514.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr11_+_844406 | 0.10 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr2_-_203735484 | 0.10 |
ENST00000420558.1 ENST00000418208.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr5_+_72921983 | 0.10 |
ENST00000296794.6 ENST00000545377.1 ENST00000513042.2 ENST00000287898.5 ENST00000509848.1 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr2_+_170440902 | 0.10 |
ENST00000448752.2 ENST00000418888.1 ENST00000414307.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr7_+_28452130 | 0.10 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr12_-_110511424 | 0.10 |
ENST00000548191.1 |
C12orf76 |
chromosome 12 open reading frame 76 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 2.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1904172 | NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:1904139 | negative regulation of norepinephrine secretion(GO:0010700) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0021930 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 2.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0004320 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |