Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for GATA5
Z-value: 0.25
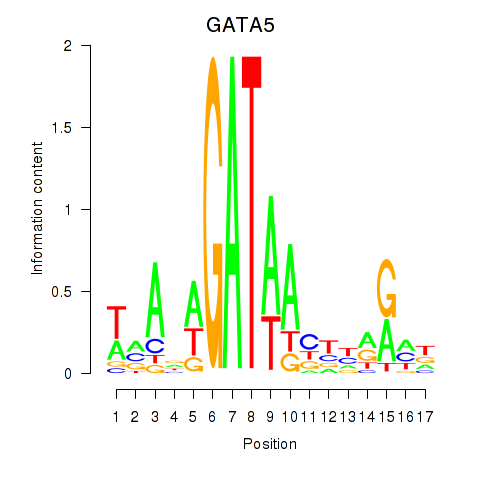
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.6 | GATA5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg19_v2_chr20_-_61051026_61051057 | 0.46 | 2.5e-01 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_118187379 | 0.56 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr21_+_25801041 | 0.36 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr2_+_102615416 | 0.31 |
ENST00000393414.2 |
IL1R2 |
interleukin 1 receptor, type II |
| chr7_+_26332645 | 0.24 |
ENST00000396376.1 |
SNX10 |
sorting nexin 10 |
| chr5_+_140227048 | 0.24 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr14_+_61995722 | 0.22 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr1_+_62439037 | 0.22 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr14_-_61748550 | 0.22 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr5_+_140261703 | 0.20 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr4_-_72649763 | 0.20 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr17_-_41277467 | 0.18 |
ENST00000494123.1 ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1 |
breast cancer 1, early onset |
| chr17_+_1633755 | 0.17 |
ENST00000545662.1 |
WDR81 |
WD repeat domain 81 |
| chr11_+_119205222 | 0.16 |
ENST00000311413.4 |
RNF26 |
ring finger protein 26 |
| chrX_+_30261847 | 0.15 |
ENST00000378981.3 ENST00000397550.1 |
MAGEB1 |
melanoma antigen family B, 1 |
| chr1_-_247921982 | 0.15 |
ENST00000408896.2 |
OR1C1 |
olfactory receptor, family 1, subfamily C, member 1 |
| chrX_-_117119243 | 0.14 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr4_-_48782259 | 0.13 |
ENST00000507711.1 ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL |
FRY-like |
| chr1_-_157014865 | 0.13 |
ENST00000361409.2 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr15_-_63674034 | 0.13 |
ENST00000344366.3 ENST00000422263.2 |
CA12 |
carbonic anhydrase XII |
| chr4_+_155484155 | 0.13 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr5_+_145316120 | 0.13 |
ENST00000359120.4 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr11_-_11747257 | 0.12 |
ENST00000601641.1 |
AC131935.1 |
AC131935.1 |
| chr14_+_45605157 | 0.12 |
ENST00000542564.2 |
FANCM |
Fanconi anemia, complementation group M |
| chr4_-_110723194 | 0.12 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr15_-_63674218 | 0.12 |
ENST00000178638.3 |
CA12 |
carbonic anhydrase XII |
| chr11_+_128563652 | 0.11 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_16793160 | 0.11 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr4_-_110723134 | 0.11 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr4_-_110723335 | 0.11 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr8_-_91095099 | 0.11 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr3_-_197300194 | 0.11 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr11_+_124824000 | 0.11 |
ENST00000529051.1 ENST00000344762.5 |
CCDC15 |
coiled-coil domain containing 15 |
| chr12_+_20968608 | 0.10 |
ENST00000381541.3 ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3 SLCO1B3 SLCO1B7 |
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chrX_-_13338518 | 0.10 |
ENST00000380622.2 |
ATXN3L |
ataxin 3-like |
| chr16_-_87466740 | 0.10 |
ENST00000561928.1 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
| chr1_+_24645865 | 0.10 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_115572415 | 0.10 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chrX_-_70326455 | 0.10 |
ENST00000374251.5 |
CXorf65 |
chromosome X open reading frame 65 |
| chr19_-_14889349 | 0.10 |
ENST00000315576.3 ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr2_+_69201705 | 0.10 |
ENST00000377938.2 |
GKN1 |
gastrokine 1 |
| chr1_+_24645807 | 0.10 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr9_-_6645628 | 0.09 |
ENST00000321612.6 |
GLDC |
glycine dehydrogenase (decarboxylating) |
| chr19_+_42580274 | 0.09 |
ENST00000359044.4 |
ZNF574 |
zinc finger protein 574 |
| chr13_+_108870714 | 0.09 |
ENST00000375898.3 |
ABHD13 |
abhydrolase domain containing 13 |
| chr6_-_32908765 | 0.09 |
ENST00000416244.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr7_+_136553370 | 0.09 |
ENST00000445907.2 |
CHRM2 |
cholinergic receptor, muscarinic 2 |
| chr5_+_147443534 | 0.09 |
ENST00000398454.1 ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5 |
serine peptidase inhibitor, Kazal type 5 |
| chr13_-_46756351 | 0.09 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_-_55739542 | 0.09 |
ENST00000446683.2 |
BMP5 |
bone morphogenetic protein 5 |
| chrX_+_105192423 | 0.09 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr8_+_9046503 | 0.08 |
ENST00000512942.2 |
RP11-10A14.5 |
RP11-10A14.5 |
| chr4_-_159956333 | 0.08 |
ENST00000434826.2 |
C4orf45 |
chromosome 4 open reading frame 45 |
| chr13_-_99910673 | 0.08 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr3_+_118905564 | 0.08 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr3_+_46283916 | 0.08 |
ENST00000395940.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr22_-_29107919 | 0.08 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr3_+_46283863 | 0.08 |
ENST00000545097.1 ENST00000541018.1 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr12_+_9102632 | 0.08 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr4_-_100575781 | 0.08 |
ENST00000511828.1 |
RP11-766F14.2 |
Protein LOC285556 |
| chr6_-_90529418 | 0.08 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr16_+_31539183 | 0.08 |
ENST00000302312.4 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr1_+_196857144 | 0.08 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr1_+_171454639 | 0.08 |
ENST00000392078.3 ENST00000426496.2 |
PRRC2C |
proline-rich coiled-coil 2C |
| chrX_-_65253506 | 0.08 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr1_+_156123318 | 0.08 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr13_+_32313658 | 0.07 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr20_+_44258165 | 0.07 |
ENST00000372643.3 |
WFDC10A |
WAP four-disulfide core domain 10A |
| chr15_+_66874502 | 0.07 |
ENST00000558797.1 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
| chr14_+_45605127 | 0.07 |
ENST00000556036.1 ENST00000267430.5 |
FANCM |
Fanconi anemia, complementation group M |
| chr8_-_6875778 | 0.07 |
ENST00000535841.1 ENST00000327857.2 |
DEFA1B DEFA3 |
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr6_-_154677900 | 0.07 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr8_-_6837602 | 0.07 |
ENST00000382692.2 |
DEFA1 |
defensin, alpha 1 |
| chr1_+_171454659 | 0.07 |
ENST00000367742.3 ENST00000338920.4 |
PRRC2C |
proline-rich coiled-coil 2C |
| chr1_-_160681593 | 0.07 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr2_+_29336855 | 0.07 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr11_-_7847519 | 0.07 |
ENST00000328375.1 |
OR5P3 |
olfactory receptor, family 5, subfamily P, member 3 |
| chr10_-_98031265 | 0.07 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr5_+_179125907 | 0.07 |
ENST00000247461.4 ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX |
calnexin |
| chr5_-_58652788 | 0.07 |
ENST00000405755.2 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr17_+_7308172 | 0.07 |
ENST00000575301.1 |
NLGN2 |
neuroligin 2 |
| chr15_-_34628951 | 0.06 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chrX_-_72097698 | 0.06 |
ENST00000373530.1 |
DMRTC1 |
DMRT-like family C1 |
| chr11_+_121461097 | 0.06 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr19_+_9296279 | 0.06 |
ENST00000344248.2 |
OR7D2 |
olfactory receptor, family 7, subfamily D, member 2 |
| chrM_+_10758 | 0.06 |
ENST00000361381.2 |
MT-ND4 |
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_+_101659682 | 0.06 |
ENST00000465215.1 |
RP11-221J22.1 |
RP11-221J22.1 |
| chr5_+_179125368 | 0.06 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr21_-_31864275 | 0.06 |
ENST00000334063.4 |
KRTAP19-3 |
keratin associated protein 19-3 |
| chr1_+_57320437 | 0.06 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr1_-_27481401 | 0.05 |
ENST00000263980.3 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr3_+_57881966 | 0.05 |
ENST00000495364.1 |
SLMAP |
sarcolemma associated protein |
| chr1_-_100643765 | 0.05 |
ENST00000370137.1 ENST00000370138.1 ENST00000342895.3 |
LRRC39 |
leucine rich repeat containing 39 |
| chr16_+_31366455 | 0.05 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr16_+_31366536 | 0.05 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_+_182850551 | 0.05 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr6_+_31783291 | 0.05 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr12_-_30887948 | 0.05 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr10_+_103911926 | 0.05 |
ENST00000605788.1 ENST00000405356.1 |
NOLC1 |
nucleolar and coiled-body phosphoprotein 1 |
| chr6_-_53409890 | 0.05 |
ENST00000229416.6 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
| chr15_-_54025300 | 0.05 |
ENST00000559418.1 |
WDR72 |
WD repeat domain 72 |
| chr6_+_160327974 | 0.05 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr12_-_9102549 | 0.05 |
ENST00000000412.3 |
M6PR |
mannose-6-phosphate receptor (cation dependent) |
| chr1_-_113478603 | 0.05 |
ENST00000443580.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr3_+_193310918 | 0.05 |
ENST00000361908.3 ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1 |
optic atrophy 1 (autosomal dominant) |
| chr1_+_196743943 | 0.05 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr14_-_24553834 | 0.05 |
ENST00000397002.2 |
NRL |
neural retina leucine zipper |
| chr19_+_7030589 | 0.05 |
ENST00000329753.5 |
MBD3L5 |
methyl-CpG binding domain protein 3-like 5 |
| chr18_+_21032781 | 0.05 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr7_+_77469439 | 0.04 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr10_+_62538089 | 0.04 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr19_-_7040190 | 0.04 |
ENST00000381394.4 |
MBD3L4 |
methyl-CpG binding domain protein 3-like 4 |
| chr8_+_77318769 | 0.04 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr11_+_57529234 | 0.04 |
ENST00000360682.6 ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
| chr2_-_202562774 | 0.04 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr9_+_106856831 | 0.04 |
ENST00000303219.8 ENST00000374787.3 |
SMC2 |
structural maintenance of chromosomes 2 |
| chr2_+_179149636 | 0.04 |
ENST00000409631.1 |
OSBPL6 |
oxysterol binding protein-like 6 |
| chr10_+_7745232 | 0.04 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr3_+_169755715 | 0.04 |
ENST00000355897.5 |
GPR160 |
G protein-coupled receptor 160 |
| chr13_+_107029084 | 0.04 |
ENST00000444865.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr10_+_7745303 | 0.04 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_65275162 | 0.04 |
ENST00000381210.3 ENST00000507440.1 |
TECRL |
trans-2,3-enoyl-CoA reductase-like |
| chr14_+_21785693 | 0.04 |
ENST00000382933.4 ENST00000557351.1 |
RPGRIP1 |
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr1_-_182921119 | 0.04 |
ENST00000423786.1 |
SHCBP1L |
SHC SH2-domain binding protein 1-like |
| chr14_-_82000140 | 0.04 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr1_-_27226928 | 0.04 |
ENST00000361720.5 |
GPATCH3 |
G patch domain containing 3 |
| chr2_-_202562716 | 0.04 |
ENST00000428900.2 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr20_+_22034809 | 0.04 |
ENST00000449427.1 |
RP11-125P18.1 |
RP11-125P18.1 |
| chr1_-_185597619 | 0.04 |
ENST00000608417.1 ENST00000436955.1 |
GS1-204I12.1 |
GS1-204I12.1 |
| chr5_+_140579162 | 0.04 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr8_+_27629459 | 0.04 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr7_-_81399411 | 0.04 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_+_45028719 | 0.03 |
ENST00000560442.1 ENST00000558329.1 ENST00000561043.1 |
TRIM69 |
tripartite motif containing 69 |
| chr1_-_157670647 | 0.03 |
ENST00000368184.3 |
FCRL3 |
Fc receptor-like 3 |
| chr15_+_45028753 | 0.03 |
ENST00000338264.4 |
TRIM69 |
tripartite motif containing 69 |
| chr6_-_86099898 | 0.03 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr6_+_153019023 | 0.03 |
ENST00000367245.5 ENST00000529453.1 |
MYCT1 |
myc target 1 |
| chr10_+_18689637 | 0.03 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_+_44916098 | 0.03 |
ENST00000296125.4 |
TGM4 |
transglutaminase 4 |
| chr6_-_135271219 | 0.03 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr12_+_57881740 | 0.03 |
ENST00000262027.5 ENST00000315473.5 |
MARS |
methionyl-tRNA synthetase |
| chr12_-_57882577 | 0.03 |
ENST00000393797.2 |
ARHGAP9 |
Rho GTPase activating protein 9 |
| chr19_-_14952689 | 0.03 |
ENST00000248058.1 |
OR7A10 |
olfactory receptor, family 7, subfamily A, member 10 |
| chr6_-_135271260 | 0.03 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr5_+_147774275 | 0.03 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr3_+_184080387 | 0.03 |
ENST00000455712.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr6_-_117150198 | 0.03 |
ENST00000310357.3 ENST00000368549.3 ENST00000530250.1 |
GPRC6A |
G protein-coupled receptor, family C, group 6, member A |
| chr6_+_118869452 | 0.03 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr14_+_67291158 | 0.03 |
ENST00000555456.1 |
GPHN |
gephyrin |
| chr2_+_216974020 | 0.03 |
ENST00000392132.2 ENST00000417391.1 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr8_+_20831460 | 0.03 |
ENST00000522604.1 |
RP11-421P23.1 |
RP11-421P23.1 |
| chr3_-_156878540 | 0.03 |
ENST00000461804.1 |
CCNL1 |
cyclin L1 |
| chr6_-_137365402 | 0.03 |
ENST00000541547.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr3_-_108476231 | 0.03 |
ENST00000295755.6 |
RETNLB |
resistin like beta |
| chr2_-_209054709 | 0.03 |
ENST00000449053.1 ENST00000451346.1 ENST00000341287.4 |
C2orf80 |
chromosome 2 open reading frame 80 |
| chr2_-_128642434 | 0.03 |
ENST00000393001.1 |
AMMECR1L |
AMMECR1-like |
| chr14_+_39735411 | 0.03 |
ENST00000603904.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr11_+_30252549 | 0.03 |
ENST00000254122.3 ENST00000417547.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr3_-_51813009 | 0.03 |
ENST00000398780.3 |
IQCF6 |
IQ motif containing F6 |
| chr2_-_136594740 | 0.03 |
ENST00000264162.2 |
LCT |
lactase |
| chr17_-_74639886 | 0.02 |
ENST00000156626.7 |
ST6GALNAC1 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr7_+_143771275 | 0.02 |
ENST00000408898.2 |
OR2A25 |
olfactory receptor, family 2, subfamily A, member 25 |
| chr6_-_29395509 | 0.02 |
ENST00000377147.2 |
OR11A1 |
olfactory receptor, family 11, subfamily A, member 1 |
| chr17_+_53344945 | 0.02 |
ENST00000575345.1 |
HLF |
hepatic leukemia factor |
| chr3_-_143567262 | 0.02 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chrM_+_10053 | 0.02 |
ENST00000361227.2 |
MT-ND3 |
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_+_38445641 | 0.02 |
ENST00000397210.3 ENST00000506135.1 ENST00000508131.1 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
| chr6_-_52710893 | 0.02 |
ENST00000284562.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr17_-_74547256 | 0.02 |
ENST00000589145.1 |
CYGB |
cytoglobin |
| chr2_-_40006289 | 0.02 |
ENST00000260619.6 ENST00000454352.2 |
THUMPD2 |
THUMP domain containing 2 |
| chrX_+_1733876 | 0.02 |
ENST00000381241.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr14_-_105420241 | 0.02 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr14_-_47812321 | 0.02 |
ENST00000357362.3 ENST00000486952.2 ENST00000426342.1 |
MDGA2 |
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr20_-_44333658 | 0.02 |
ENST00000330523.5 ENST00000335769.2 |
WFDC10B |
WAP four-disulfide core domain 10B |
| chr4_+_155484103 | 0.02 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr12_-_45270151 | 0.02 |
ENST00000429094.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr11_-_72070206 | 0.02 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr4_-_70518941 | 0.02 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr6_+_140175987 | 0.02 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr12_-_45269430 | 0.02 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr12_-_45270077 | 0.02 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr17_-_72772462 | 0.02 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_+_18433840 | 0.02 |
ENST00000541669.1 ENST00000280704.4 |
LDHC |
lactate dehydrogenase C |
| chr1_-_17380630 | 0.02 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chrX_+_1734051 | 0.01 |
ENST00000381229.4 ENST00000381233.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr16_+_56485402 | 0.01 |
ENST00000566157.1 ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1 |
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr7_-_86688990 | 0.01 |
ENST00000450689.2 |
KIAA1324L |
KIAA1324-like |
| chr12_-_68619586 | 0.01 |
ENST00000229134.4 |
IL26 |
interleukin 26 |
| chr13_+_27844464 | 0.01 |
ENST00000241463.4 |
RASL11A |
RAS-like, family 11, member A |
| chr6_+_32709119 | 0.01 |
ENST00000374940.3 |
HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
| chr11_+_118754475 | 0.01 |
ENST00000292174.4 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
| chr13_+_107028897 | 0.01 |
ENST00000439790.1 ENST00000435024.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr1_+_10292308 | 0.01 |
ENST00000377081.1 |
KIF1B |
kinesin family member 1B |
| chrX_+_11777671 | 0.01 |
ENST00000380693.3 ENST00000380692.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr12_+_75874580 | 0.01 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_-_9268707 | 0.01 |
ENST00000318602.7 |
A2M |
alpha-2-macroglobulin |
| chr2_-_47143160 | 0.01 |
ENST00000409800.1 ENST00000409218.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr1_+_150954493 | 0.01 |
ENST00000368947.4 |
ANXA9 |
annexin A9 |
| chrX_+_30260054 | 0.01 |
ENST00000378982.2 |
MAGEB4 |
melanoma antigen family B, 4 |
| chr19_+_35820064 | 0.01 |
ENST00000341773.6 ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22 |
CD22 molecule |
| chrX_-_100129128 | 0.01 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |