Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for GFI1B
Z-value: 0.52
Transcription factors associated with GFI1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1B
|
ENSG00000165702.8 | GFI1B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1B | hg19_v2_chr9_+_135854091_135854159 | -0.66 | 7.3e-02 | Click! |
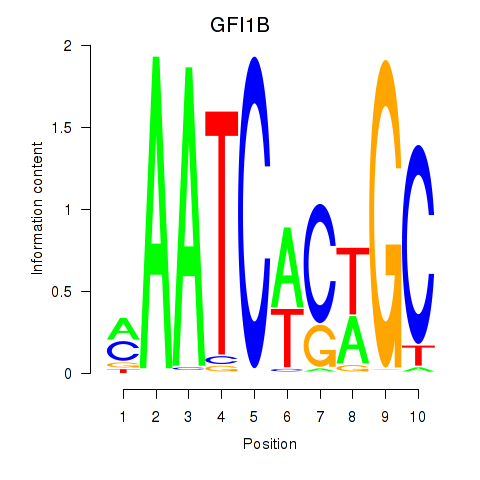
Activity profile of GFI1B motif
Sorted Z-values of GFI1B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31610064 | 0.40 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr2_+_152214098 | 0.35 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr13_+_76362974 | 0.32 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr6_-_56707943 | 0.30 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr14_-_53417732 | 0.29 |
ENST00000399304.3 ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2 |
fermitin family member 2 |
| chr16_+_12995614 | 0.26 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr1_+_210406121 | 0.24 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr13_-_67802549 | 0.22 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr6_-_56708459 | 0.19 |
ENST00000370788.2 |
DST |
dystonin |
| chr12_+_117176090 | 0.19 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr14_-_80677815 | 0.19 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr12_+_117176113 | 0.19 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr10_-_49813090 | 0.19 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr5_+_98109322 | 0.18 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr2_-_190044480 | 0.18 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr5_-_124080203 | 0.18 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr19_+_36602104 | 0.17 |
ENST00000585332.1 ENST00000262637.4 |
OVOL3 |
ovo-like zinc finger 3 |
| chr16_+_12995468 | 0.16 |
ENST00000424107.3 ENST00000558583.1 ENST00000558318.1 |
SHISA9 |
shisa family member 9 |
| chr1_+_162602244 | 0.16 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr12_-_53601055 | 0.15 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr12_-_53601000 | 0.15 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr14_+_29236269 | 0.14 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr6_-_31648150 | 0.14 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr12_+_51818586 | 0.14 |
ENST00000394856.1 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr12_+_51818749 | 0.14 |
ENST00000514353.3 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr16_+_88493879 | 0.13 |
ENST00000565624.1 ENST00000437464.1 |
ZNF469 |
zinc finger protein 469 |
| chr20_+_32254286 | 0.13 |
ENST00000330271.4 |
ACTL10 |
actin-like 10 |
| chr1_+_201755452 | 0.13 |
ENST00000438083.1 |
NAV1 |
neuron navigator 1 |
| chr10_+_17272608 | 0.12 |
ENST00000421459.2 |
VIM |
vimentin |
| chr21_-_42879909 | 0.12 |
ENST00000458356.1 ENST00000398585.3 ENST00000424093.1 |
TMPRSS2 |
transmembrane protease, serine 2 |
| chr7_-_138363824 | 0.11 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr4_+_160188889 | 0.11 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_179834311 | 0.11 |
ENST00000553856.1 |
IFRG15 |
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr12_+_51818555 | 0.11 |
ENST00000453097.2 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr10_-_129924611 | 0.11 |
ENST00000368654.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr10_-_129924468 | 0.11 |
ENST00000368653.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr3_+_149689066 | 0.11 |
ENST00000593416.1 |
AC117395.1 |
LOC646903 protein; Uncharacterized protein |
| chr11_-_61582579 | 0.11 |
ENST00000539419.1 ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1 |
fatty acid desaturase 1 |
| chr13_+_113623509 | 0.10 |
ENST00000535094.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chrX_+_7137475 | 0.10 |
ENST00000217961.4 |
STS |
steroid sulfatase (microsomal), isozyme S |
| chr22_+_19467261 | 0.10 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chr17_-_5321549 | 0.10 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr15_+_32933866 | 0.10 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr2_-_45236540 | 0.10 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr16_+_30996502 | 0.10 |
ENST00000353250.5 ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_54683419 | 0.10 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr17_+_74075263 | 0.10 |
ENST00000334586.5 ENST00000392503.2 |
ZACN |
zinc activated ligand-gated ion channel |
| chr2_-_127963343 | 0.10 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr9_-_123239632 | 0.09 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr12_+_53835508 | 0.09 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr1_+_164528866 | 0.09 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr12_+_117348742 | 0.09 |
ENST00000309909.5 ENST00000455858.2 |
FBXW8 |
F-box and WD repeat domain containing 8 |
| chr6_-_13621126 | 0.09 |
ENST00000600057.1 |
AL441883.1 |
Uncharacterized protein |
| chrX_+_57618269 | 0.09 |
ENST00000374888.1 |
ZXDB |
zinc finger, X-linked, duplicated B |
| chr17_-_33864772 | 0.09 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr3_+_152017924 | 0.09 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr22_+_38864041 | 0.09 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_+_204732666 | 0.08 |
ENST00000295854.6 ENST00000472206.1 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr7_-_14029515 | 0.08 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr6_+_12290586 | 0.08 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr5_-_64920115 | 0.08 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr11_-_64703354 | 0.08 |
ENST00000532246.1 ENST00000279168.2 |
GPHA2 |
glycoprotein hormone alpha 2 |
| chr15_+_38544476 | 0.08 |
ENST00000299084.4 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
| chr7_+_134576317 | 0.08 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr1_+_182419261 | 0.08 |
ENST00000294854.8 ENST00000542961.1 |
RGSL1 |
regulator of G-protein signaling like 1 |
| chr5_+_140529630 | 0.08 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr10_+_60936921 | 0.08 |
ENST00000373878.3 |
PHYHIPL |
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr9_-_123638633 | 0.08 |
ENST00000456291.1 |
PHF19 |
PHD finger protein 19 |
| chr8_-_102275131 | 0.07 |
ENST00000523121.1 |
KB-1410C5.2 |
KB-1410C5.2 |
| chr4_-_104640973 | 0.07 |
ENST00000304883.2 |
TACR3 |
tachykinin receptor 3 |
| chrX_+_49832231 | 0.07 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr1_-_182360498 | 0.07 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr1_+_203651937 | 0.07 |
ENST00000341360.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr9_-_110540419 | 0.07 |
ENST00000398726.3 |
AL162389.1 |
Uncharacterized protein |
| chr12_-_82153087 | 0.07 |
ENST00000547623.1 ENST00000549396.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_+_109102652 | 0.07 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr17_-_47287928 | 0.07 |
ENST00000507680.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr18_+_77905794 | 0.07 |
ENST00000587254.1 ENST00000586421.1 |
AC139100.2 |
Uncharacterized protein |
| chr6_-_26108355 | 0.07 |
ENST00000338379.4 |
HIST1H1T |
histone cluster 1, H1t |
| chr1_-_92764523 | 0.07 |
ENST00000370360.3 ENST00000534881.1 |
GLMN |
glomulin, FKBP associated protein |
| chr17_-_33759509 | 0.07 |
ENST00000304905.5 |
SLFN12 |
schlafen family member 12 |
| chr16_-_18441131 | 0.07 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr17_-_50236039 | 0.07 |
ENST00000451037.2 |
CA10 |
carbonic anhydrase X |
| chr1_-_68698222 | 0.07 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr10_+_94590910 | 0.07 |
ENST00000371547.4 |
EXOC6 |
exocyst complex component 6 |
| chr5_-_139930713 | 0.07 |
ENST00000602657.1 |
SRA1 |
steroid receptor RNA activator 1 |
| chr5_+_14143728 | 0.07 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr17_-_295730 | 0.07 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr22_+_22020273 | 0.06 |
ENST00000412327.1 ENST00000335025.8 ENST00000398831.3 ENST00000492445.2 ENST00000458567.1 ENST00000406385.1 |
PPIL2 |
peptidylprolyl isomerase (cyclophilin)-like 2 |
| chr14_-_80677970 | 0.06 |
ENST00000438257.4 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr12_+_72332618 | 0.06 |
ENST00000333850.3 |
TPH2 |
tryptophan hydroxylase 2 |
| chr18_+_48494361 | 0.06 |
ENST00000588577.1 ENST00000269466.3 ENST00000591429.1 ENST00000452201.2 |
ELAC1 SMAD4 |
elaC ribonuclease Z 1 SMAD family member 4 |
| chr7_+_150758304 | 0.06 |
ENST00000482950.1 ENST00000463414.1 ENST00000310317.5 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr15_+_71228826 | 0.06 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr1_-_182361327 | 0.06 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr20_-_35274548 | 0.06 |
ENST00000262866.4 |
SLA2 |
Src-like-adaptor 2 |
| chr19_+_21264980 | 0.06 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr10_-_102279586 | 0.06 |
ENST00000370345.3 ENST00000451524.1 ENST00000370329.5 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
| chr19_+_55417530 | 0.06 |
ENST00000350790.5 ENST00000338835.5 ENST00000357397.5 |
NCR1 |
natural cytotoxicity triggering receptor 1 |
| chr1_+_39796810 | 0.06 |
ENST00000289893.4 |
MACF1 |
microtubule-actin crosslinking factor 1 |
| chr1_-_182360918 | 0.06 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr4_+_26585538 | 0.06 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr1_+_13359819 | 0.06 |
ENST00000376168.1 |
PRAMEF5 |
PRAME family member 5 |
| chr7_+_120591170 | 0.06 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr17_-_1395954 | 0.06 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chr5_+_64920543 | 0.06 |
ENST00000399438.3 ENST00000510585.2 |
TRAPPC13 CTC-534A2.2 |
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr19_-_58951496 | 0.06 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr19_+_55327923 | 0.06 |
ENST00000391728.4 ENST00000326542.7 ENST00000358178.4 |
KIR3DL1 |
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 |
| chr7_+_120590803 | 0.06 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr16_+_31366536 | 0.06 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chrX_-_71933888 | 0.06 |
ENST00000373542.4 ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
| chr4_+_56814968 | 0.06 |
ENST00000422247.2 |
CEP135 |
centrosomal protein 135kDa |
| chr5_+_159436120 | 0.06 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr7_+_127233689 | 0.05 |
ENST00000265825.5 ENST00000420086.2 |
FSCN3 |
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr11_-_102595681 | 0.05 |
ENST00000236826.3 |
MMP8 |
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr1_-_68698197 | 0.05 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_-_9714644 | 0.05 |
ENST00000377320.3 |
C1orf200 |
chromosome 1 open reading frame 200 |
| chr11_-_82745238 | 0.05 |
ENST00000531021.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr7_+_134576151 | 0.05 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr1_-_13002348 | 0.05 |
ENST00000355096.2 |
PRAMEF6 |
PRAME family member 6 |
| chr14_+_105957402 | 0.05 |
ENST00000421892.1 ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80 |
chromosome 14 open reading frame 80 |
| chr1_-_13007420 | 0.05 |
ENST00000376189.1 |
PRAMEF6 |
PRAME family member 6 |
| chr12_+_100967420 | 0.05 |
ENST00000266754.5 ENST00000547754.1 |
GAS2L3 |
growth arrest-specific 2 like 3 |
| chr12_-_4553385 | 0.05 |
ENST00000543077.1 |
FGF6 |
fibroblast growth factor 6 |
| chr2_-_10978103 | 0.05 |
ENST00000404824.2 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr17_+_8213590 | 0.05 |
ENST00000361926.3 |
ARHGEF15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr11_-_5462744 | 0.05 |
ENST00000380211.1 |
OR51I1 |
olfactory receptor, family 51, subfamily I, member 1 |
| chr2_-_27938593 | 0.05 |
ENST00000379677.2 |
AC074091.13 |
Uncharacterized protein |
| chr16_+_50775971 | 0.05 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr3_+_131100515 | 0.05 |
ENST00000537561.1 ENST00000359850.3 ENST00000521288.1 ENST00000502852.1 |
NUDT16 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr16_+_68279207 | 0.05 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr7_+_134551583 | 0.05 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr19_-_51412584 | 0.05 |
ENST00000431178.2 |
KLK4 |
kallikrein-related peptidase 4 |
| chr2_+_69201705 | 0.05 |
ENST00000377938.2 |
GKN1 |
gastrokine 1 |
| chr19_+_58258164 | 0.05 |
ENST00000317178.5 |
ZNF776 |
zinc finger protein 776 |
| chr2_-_158732340 | 0.05 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr15_-_90456156 | 0.04 |
ENST00000357484.5 |
C15orf38 |
chromosome 15 open reading frame 38 |
| chr19_+_10736183 | 0.04 |
ENST00000590857.1 ENST00000588688.1 ENST00000586078.1 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
| chr2_+_233390863 | 0.04 |
ENST00000449596.1 ENST00000543200.1 |
CHRND |
cholinergic receptor, nicotinic, delta (muscle) |
| chrX_+_46696372 | 0.04 |
ENST00000218340.3 |
RP2 |
retinitis pigmentosa 2 (X-linked recessive) |
| chr16_-_18468926 | 0.04 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_25963020 | 0.04 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr17_-_79481666 | 0.04 |
ENST00000575659.1 |
ACTG1 |
actin, gamma 1 |
| chr14_+_77228532 | 0.04 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr3_+_152017360 | 0.04 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr1_-_220219775 | 0.04 |
ENST00000609181.1 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr19_-_46088068 | 0.04 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr16_-_30597000 | 0.04 |
ENST00000470110.1 ENST00000395216.2 |
ZNF785 |
zinc finger protein 785 |
| chr9_+_35538616 | 0.04 |
ENST00000455600.1 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr17_-_79827808 | 0.04 |
ENST00000580685.1 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr5_+_109025067 | 0.04 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr19_-_35719609 | 0.04 |
ENST00000324675.3 |
FAM187B |
family with sequence similarity 187, member B |
| chr20_-_33999766 | 0.04 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr1_-_13005246 | 0.04 |
ENST00000415464.2 |
PRAMEF6 |
PRAME family member 6 |
| chr16_+_31366455 | 0.04 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_-_85788605 | 0.04 |
ENST00000233838.4 |
GGCX |
gamma-glutamyl carboxylase |
| chr6_-_131384347 | 0.04 |
ENST00000530481.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr12_+_6603253 | 0.04 |
ENST00000382457.4 ENST00000545962.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr22_+_19466980 | 0.04 |
ENST00000407835.1 ENST00000438587.1 |
CDC45 |
cell division cycle 45 |
| chr18_-_5419797 | 0.04 |
ENST00000542146.1 ENST00000427684.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr4_+_56815102 | 0.04 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr21_+_35014783 | 0.04 |
ENST00000381291.4 ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr11_-_7961141 | 0.04 |
ENST00000360759.3 |
OR10A3 |
olfactory receptor, family 10, subfamily A, member 3 |
| chr6_-_131321863 | 0.04 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr10_+_63661053 | 0.04 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr6_-_131384373 | 0.04 |
ENST00000392427.3 ENST00000525271.1 ENST00000527411.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr17_+_61554413 | 0.04 |
ENST00000538928.1 ENST00000290866.4 ENST00000428043.1 |
ACE |
angiotensin I converting enzyme |
| chr11_+_119056178 | 0.04 |
ENST00000525131.1 ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3 |
PDZ domain containing 3 |
| chr5_-_125930929 | 0.04 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr1_-_230513367 | 0.04 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr18_-_30050395 | 0.04 |
ENST00000269209.6 ENST00000399218.4 |
GAREM |
GRB2 associated, regulator of MAPK1 |
| chr1_-_47134085 | 0.04 |
ENST00000371937.4 ENST00000574428.1 ENST00000329231.4 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr7_-_27196267 | 0.04 |
ENST00000242159.3 |
HOXA7 |
homeobox A7 |
| chr12_+_96337061 | 0.04 |
ENST00000266736.2 |
AMDHD1 |
amidohydrolase domain containing 1 |
| chr16_+_50775948 | 0.04 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr14_-_68162464 | 0.04 |
ENST00000553384.1 ENST00000557726.1 ENST00000381346.4 |
RDH11 |
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr13_-_27745936 | 0.04 |
ENST00000282344.6 |
USP12 |
ubiquitin specific peptidase 12 |
| chr10_+_114206956 | 0.04 |
ENST00000432306.1 ENST00000393077.2 |
VTI1A |
vesicle transport through interaction with t-SNAREs 1A |
| chr3_-_158450231 | 0.04 |
ENST00000479756.1 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr2_+_201994042 | 0.04 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr11_-_68780824 | 0.04 |
ENST00000441623.1 ENST00000309099.6 |
MRGPRF |
MAS-related GPR, member F |
| chr9_+_35490101 | 0.04 |
ENST00000361226.3 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr4_+_78829479 | 0.04 |
ENST00000504901.1 |
MRPL1 |
mitochondrial ribosomal protein L1 |
| chr1_+_176432298 | 0.04 |
ENST00000367661.3 ENST00000367662.3 |
PAPPA2 |
pappalysin 2 |
| chr7_-_100808394 | 0.03 |
ENST00000445482.2 |
VGF |
VGF nerve growth factor inducible |
| chr1_-_47134101 | 0.03 |
ENST00000576409.1 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr11_+_101785727 | 0.03 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr6_-_131384412 | 0.03 |
ENST00000445890.2 ENST00000368128.2 ENST00000337057.3 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr10_+_52152766 | 0.03 |
ENST00000596442.1 |
AC069547.2 |
Uncharacterized protein |
| chr8_+_24298597 | 0.03 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr6_+_26521948 | 0.03 |
ENST00000411553.1 |
HCG11 |
HLA complex group 11 (non-protein coding) |
| chr6_-_76782371 | 0.03 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chrX_+_129473916 | 0.03 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_201994208 | 0.03 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_+_202122703 | 0.03 |
ENST00000447616.1 ENST00000358485.4 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
| chr12_-_118406028 | 0.03 |
ENST00000425217.1 |
KSR2 |
kinase suppressor of ras 2 |
| chr2_-_85788652 | 0.03 |
ENST00000430215.3 |
GGCX |
gamma-glutamyl carboxylase |
| chr21_+_45705752 | 0.03 |
ENST00000291582.5 |
AIRE |
autoimmune regulator |
| chrX_-_103268259 | 0.03 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chr15_-_90645679 | 0.03 |
ENST00000539790.1 ENST00000559482.1 ENST00000330062.3 |
IDH2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.0 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.0 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0000799 | nuclear condensin complex(GO:0000799) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |