Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
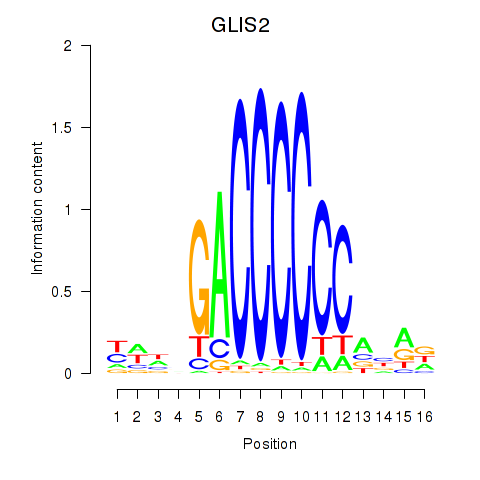
Results for GLIS2
Z-value: 0.75
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4364762_4364762, hg19_v2_chr16_+_4382225_4382225 | -0.10 | 8.1e-01 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_44637526 | 0.76 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr14_-_105635090 | 0.76 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr19_-_6720686 | 0.55 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr5_+_68788594 | 0.50 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr1_-_28520447 | 0.49 |
ENST00000539896.1 |
PTAFR |
platelet-activating factor receptor |
| chr12_-_51785182 | 0.45 |
ENST00000356317.3 ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr7_+_69064300 | 0.44 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr16_-_68269971 | 0.42 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr5_-_176836577 | 0.40 |
ENST00000253496.3 |
F12 |
coagulation factor XII (Hageman factor) |
| chr13_-_20767037 | 0.37 |
ENST00000382848.4 |
GJB2 |
gap junction protein, beta 2, 26kDa |
| chr2_+_220492373 | 0.36 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr1_-_186649543 | 0.36 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_-_55652290 | 0.35 |
ENST00000589745.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr1_-_156675535 | 0.35 |
ENST00000368221.1 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr19_+_45409011 | 0.34 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr3_-_48470838 | 0.34 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr18_+_11752040 | 0.33 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr19_-_49567124 | 0.32 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr1_-_209824643 | 0.31 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr1_+_65613217 | 0.31 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr1_-_156675368 | 0.31 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr19_+_17858509 | 0.29 |
ENST00000594202.1 ENST00000252771.7 ENST00000389133.4 |
FCHO1 |
FCH domain only 1 |
| chr20_+_30640004 | 0.29 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr22_-_39640756 | 0.27 |
ENST00000331163.6 |
PDGFB |
platelet-derived growth factor beta polypeptide |
| chr22_+_45072925 | 0.27 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr8_+_98881268 | 0.27 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr22_+_45072958 | 0.27 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr14_+_94640633 | 0.26 |
ENST00000304338.3 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
| chr11_+_64058758 | 0.25 |
ENST00000538767.1 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr11_-_72353451 | 0.24 |
ENST00000376450.3 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr14_-_92302784 | 0.24 |
ENST00000340892.5 ENST00000360594.5 |
TC2N |
tandem C2 domains, nuclear |
| chr17_+_78075361 | 0.24 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr19_+_49891475 | 0.23 |
ENST00000447857.3 |
CCDC155 |
coiled-coil domain containing 155 |
| chr16_-_29910365 | 0.23 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr19_+_45417504 | 0.22 |
ENST00000588750.1 ENST00000588802.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_45417812 | 0.22 |
ENST00000592535.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_17858547 | 0.22 |
ENST00000600676.1 ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1 |
FCH domain only 1 |
| chr2_+_114737472 | 0.21 |
ENST00000420161.1 |
AC110769.3 |
AC110769.3 |
| chr6_+_33048222 | 0.21 |
ENST00000428835.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr11_+_121447469 | 0.21 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr20_+_30639991 | 0.21 |
ENST00000534862.1 ENST00000538448.1 ENST00000375862.2 |
HCK |
hemopoietic cell kinase |
| chr17_+_47572647 | 0.21 |
ENST00000172229.3 |
NGFR |
nerve growth factor receptor |
| chr19_-_50979981 | 0.20 |
ENST00000595790.1 ENST00000600100.1 |
FAM71E1 |
family with sequence similarity 71, member E1 |
| chr14_-_21566731 | 0.20 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr17_+_48503519 | 0.20 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr19_+_45418067 | 0.20 |
ENST00000589078.1 ENST00000586638.1 |
APOC1 |
apolipoprotein C-I |
| chr19_+_20959098 | 0.19 |
ENST00000360204.5 ENST00000594534.1 |
ZNF66 |
zinc finger protein 66 |
| chr17_+_48503603 | 0.19 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr13_-_114018400 | 0.19 |
ENST00000375430.4 ENST00000375431.4 |
GRTP1 |
growth hormone regulated TBC protein 1 |
| chrX_+_102469997 | 0.18 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr17_-_34257771 | 0.18 |
ENST00000394529.3 ENST00000293273.6 |
RDM1 |
RAD52 motif 1 |
| chr19_+_45417921 | 0.18 |
ENST00000252491.4 ENST00000592885.1 ENST00000589781.1 |
APOC1 |
apolipoprotein C-I |
| chr1_-_6550625 | 0.18 |
ENST00000377725.1 ENST00000340850.5 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr19_+_45349630 | 0.18 |
ENST00000252483.5 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr2_+_37571717 | 0.17 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr3_-_128840604 | 0.17 |
ENST00000476465.1 ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43 |
RAB43, member RAS oncogene family |
| chr1_-_43751230 | 0.17 |
ENST00000523677.1 |
C1orf210 |
chromosome 1 open reading frame 210 |
| chr6_-_2903514 | 0.16 |
ENST00000380698.4 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
| chr22_+_29469100 | 0.16 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr12_+_50017327 | 0.16 |
ENST00000261897.1 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr12_+_50017184 | 0.16 |
ENST00000548825.2 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr19_-_46405861 | 0.15 |
ENST00000322217.5 |
MYPOP |
Myb-related transcription factor, partner of profilin |
| chr19_-_2050852 | 0.15 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr17_-_34257731 | 0.15 |
ENST00000431884.2 ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1 |
RAD52 motif 1 |
| chr17_-_4463856 | 0.14 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr1_-_23810664 | 0.14 |
ENST00000336689.3 ENST00000437606.2 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr8_+_118533049 | 0.14 |
ENST00000522839.1 |
MED30 |
mediator complex subunit 30 |
| chr11_+_63753883 | 0.14 |
ENST00000538426.1 ENST00000543004.1 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
| chr6_-_133119668 | 0.14 |
ENST00000275227.4 ENST00000538764.1 |
SLC18B1 |
solute carrier family 18, subfamily B, member 1 |
| chr12_+_50451462 | 0.13 |
ENST00000447966.2 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr5_-_1524015 | 0.13 |
ENST00000283415.3 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
| chr11_+_68451943 | 0.13 |
ENST00000265643.3 |
GAL |
galanin/GMAP prepropeptide |
| chr10_-_14880566 | 0.13 |
ENST00000378442.1 |
CDNF |
cerebral dopamine neurotrophic factor |
| chr19_-_16008880 | 0.13 |
ENST00000011989.7 ENST00000221700.6 |
CYP4F2 |
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr19_+_2249308 | 0.13 |
ENST00000592877.1 ENST00000221496.4 |
AMH |
anti-Mullerian hormone |
| chr2_-_46769694 | 0.13 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr3_+_118892362 | 0.12 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr3_+_44626446 | 0.12 |
ENST00000441021.1 ENST00000322734.2 |
ZNF660 |
zinc finger protein 660 |
| chr2_+_11752379 | 0.12 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr17_-_34079897 | 0.12 |
ENST00000254466.6 ENST00000587565.1 |
GAS2L2 |
growth arrest-specific 2 like 2 |
| chr14_-_21493884 | 0.12 |
ENST00000556974.1 ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2 |
NDRG family member 2 |
| chr6_+_90604188 | 0.12 |
ENST00000369352.1 |
GJA10 |
gap junction protein, alpha 10, 62kDa |
| chr2_+_37571845 | 0.12 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr1_-_153348067 | 0.12 |
ENST00000368737.3 |
S100A12 |
S100 calcium binding protein A12 |
| chr19_-_42759300 | 0.12 |
ENST00000222329.4 |
ERF |
Ets2 repressor factor |
| chr16_-_32688053 | 0.12 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr19_+_14640372 | 0.11 |
ENST00000215567.5 ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR |
trans-2,3-enoyl-CoA reductase |
| chr14_-_105531759 | 0.11 |
ENST00000329797.3 ENST00000539291.2 ENST00000392585.2 |
GPR132 |
G protein-coupled receptor 132 |
| chr2_-_230933709 | 0.11 |
ENST00000436869.1 ENST00000295190.4 |
SLC16A14 |
solute carrier family 16, member 14 |
| chr17_-_41132088 | 0.11 |
ENST00000591916.1 ENST00000451885.2 ENST00000454303.1 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr16_+_2521500 | 0.11 |
ENST00000293973.1 |
NTN3 |
netrin 3 |
| chr13_+_47127293 | 0.11 |
ENST00000311191.6 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr12_+_50451331 | 0.11 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr16_+_32264040 | 0.10 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chr6_+_138725343 | 0.10 |
ENST00000607197.1 ENST00000367697.3 |
HEBP2 |
heme binding protein 2 |
| chr6_-_117086873 | 0.10 |
ENST00000368557.4 |
FAM162B |
family with sequence similarity 162, member B |
| chr17_-_7307358 | 0.10 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_-_1356628 | 0.10 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr21_+_34775698 | 0.10 |
ENST00000381995.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr16_+_9185450 | 0.10 |
ENST00000327827.7 |
C16orf72 |
chromosome 16 open reading frame 72 |
| chr12_-_53242770 | 0.10 |
ENST00000304620.4 ENST00000547110.1 |
KRT78 |
keratin 78 |
| chr3_+_52017454 | 0.09 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr7_-_155601766 | 0.09 |
ENST00000430104.1 |
SHH |
sonic hedgehog |
| chr17_+_41476327 | 0.09 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr10_+_103825080 | 0.09 |
ENST00000299238.5 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
| chrX_-_8700171 | 0.09 |
ENST00000262648.3 |
KAL1 |
Kallmann syndrome 1 sequence |
| chr3_-_125803105 | 0.09 |
ENST00000346785.5 ENST00000315891.6 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr19_+_11649532 | 0.09 |
ENST00000252456.2 ENST00000592923.1 ENST00000535659.2 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr3_-_197282821 | 0.09 |
ENST00000445160.2 ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_+_80416050 | 0.09 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr20_+_52824367 | 0.09 |
ENST00000371419.2 |
PFDN4 |
prefoldin subunit 4 |
| chr10_-_13276329 | 0.09 |
ENST00000378681.3 ENST00000463405.2 |
UCMA |
upper zone of growth plate and cartilage matrix associated |
| chr16_+_765092 | 0.09 |
ENST00000568223.2 |
METRN |
meteorin, glial cell differentiation regulator |
| chr19_-_38397285 | 0.08 |
ENST00000303868.5 |
WDR87 |
WD repeat domain 87 |
| chr16_-_4664860 | 0.08 |
ENST00000587615.1 ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1 |
UBA-like domain containing 1 |
| chr1_+_87595433 | 0.08 |
ENST00000469312.2 ENST00000490006.2 |
RP5-1052I5.1 |
long intergenic non-protein coding RNA 1140 |
| chr19_-_38397228 | 0.08 |
ENST00000447313.2 |
WDR87 |
WD repeat domain 87 |
| chr15_-_79383102 | 0.08 |
ENST00000558480.2 ENST00000419573.3 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr2_+_169312350 | 0.08 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr22_-_18257178 | 0.08 |
ENST00000342111.5 |
BID |
BH3 interacting domain death agonist |
| chr7_-_752074 | 0.08 |
ENST00000360274.4 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr16_+_27078219 | 0.08 |
ENST00000418886.1 |
C16orf82 |
chromosome 16 open reading frame 82 |
| chr1_-_40157345 | 0.08 |
ENST00000372844.3 |
HPCAL4 |
hippocalcin like 4 |
| chr19_-_2702681 | 0.08 |
ENST00000382159.3 |
GNG7 |
guanine nucleotide binding protein (G protein), gamma 7 |
| chr17_-_42200996 | 0.08 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr1_+_44412577 | 0.08 |
ENST00000372343.3 |
IPO13 |
importin 13 |
| chr19_+_17337406 | 0.08 |
ENST00000597836.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr17_-_39280419 | 0.08 |
ENST00000394014.1 |
KRTAP4-12 |
keratin associated protein 4-12 |
| chr3_+_111578131 | 0.08 |
ENST00000498699.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr19_+_45251804 | 0.08 |
ENST00000164227.5 |
BCL3 |
B-cell CLL/lymphoma 3 |
| chr21_+_42539701 | 0.08 |
ENST00000330333.6 ENST00000328735.6 ENST00000347667.5 |
BACE2 |
beta-site APP-cleaving enzyme 2 |
| chr8_-_104427313 | 0.08 |
ENST00000297578.4 |
SLC25A32 |
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr16_-_68014732 | 0.08 |
ENST00000268793.4 |
DPEP3 |
dipeptidase 3 |
| chr7_+_140373619 | 0.08 |
ENST00000483369.1 |
ADCK2 |
aarF domain containing kinase 2 |
| chr19_+_56186557 | 0.08 |
ENST00000270460.6 |
EPN1 |
epsin 1 |
| chr16_+_691792 | 0.07 |
ENST00000307650.4 |
FAM195A |
family with sequence similarity 195, member A |
| chr9_-_34710066 | 0.07 |
ENST00000378792.1 ENST00000259607.2 |
CCL21 |
chemokine (C-C motif) ligand 21 |
| chr9_+_139847347 | 0.07 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr14_-_21493649 | 0.07 |
ENST00000553442.1 ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2 |
NDRG family member 2 |
| chr9_-_98279241 | 0.07 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr17_+_64831228 | 0.07 |
ENST00000533854.1 |
CACNG5 |
calcium channel, voltage-dependent, gamma subunit 5 |
| chr13_+_47127322 | 0.07 |
ENST00000389798.3 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr4_-_48082192 | 0.07 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr19_-_3479086 | 0.07 |
ENST00000587847.1 |
C19orf77 |
chromosome 19 open reading frame 77 |
| chr16_+_33204980 | 0.07 |
ENST00000561509.1 |
TP53TG3C |
TP53 target 3C |
| chr7_+_140372953 | 0.07 |
ENST00000072869.4 ENST00000476491.1 |
ADCK2 |
aarF domain containing kinase 2 |
| chr3_+_4535025 | 0.07 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_-_27701307 | 0.07 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr7_+_29237354 | 0.07 |
ENST00000546235.1 |
CHN2 |
chimerin 2 |
| chr19_-_55669093 | 0.07 |
ENST00000344887.5 |
TNNI3 |
troponin I type 3 (cardiac) |
| chr21_-_45671014 | 0.06 |
ENST00000436357.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr7_-_130080977 | 0.06 |
ENST00000223208.5 |
CEP41 |
centrosomal protein 41kDa |
| chr19_-_19249255 | 0.06 |
ENST00000587583.2 ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A |
transmembrane protein 161A |
| chr2_+_169312725 | 0.06 |
ENST00000392687.4 |
CERS6 |
ceramide synthase 6 |
| chr22_-_30685596 | 0.06 |
ENST00000404953.3 ENST00000407689.3 |
GATSL3 |
GATS protein-like 3 |
| chr15_+_90234028 | 0.06 |
ENST00000268130.7 ENST00000560294.1 ENST00000558000.1 |
WDR93 |
WD repeat domain 93 |
| chr19_+_17337473 | 0.06 |
ENST00000598068.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr21_+_34775181 | 0.06 |
ENST00000290219.6 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr9_+_87284675 | 0.06 |
ENST00000376208.1 ENST00000304053.6 ENST00000277120.3 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr20_+_35974532 | 0.06 |
ENST00000373578.2 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr13_-_42535214 | 0.06 |
ENST00000379310.3 ENST00000281496.6 |
VWA8 |
von Willebrand factor A domain containing 8 |
| chr12_-_49110613 | 0.06 |
ENST00000261900.3 |
CCNT1 |
cyclin T1 |
| chr22_-_38794490 | 0.06 |
ENST00000400206.2 |
CSNK1E |
casein kinase 1, epsilon |
| chr2_-_131357123 | 0.06 |
ENST00000259216.4 |
CFC1 |
cripto, FRL-1, cryptic family 1 |
| chr10_+_102747783 | 0.06 |
ENST00000311916.2 ENST00000370228.1 |
C10orf2 |
chromosome 10 open reading frame 2 |
| chr3_-_193096600 | 0.06 |
ENST00000446087.1 ENST00000342358.4 |
ATP13A5 |
ATPase type 13A5 |
| chr17_-_27224621 | 0.06 |
ENST00000394906.2 ENST00000585169.1 ENST00000394908.4 |
FLOT2 |
flotillin 2 |
| chr17_+_7461580 | 0.06 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_+_45254529 | 0.05 |
ENST00000444487.1 |
BCL3 |
B-cell CLL/lymphoma 3 |
| chr2_-_85829496 | 0.05 |
ENST00000409668.1 |
TMEM150A |
transmembrane protein 150A |
| chr19_-_54784937 | 0.05 |
ENST00000434421.1 ENST00000314446.5 ENST00000391749.4 |
LILRB2 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr4_-_106394866 | 0.05 |
ENST00000502596.1 |
PPA2 |
pyrophosphatase (inorganic) 2 |
| chr7_-_98741714 | 0.05 |
ENST00000361125.1 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr17_+_7210898 | 0.05 |
ENST00000572815.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr6_-_39290744 | 0.05 |
ENST00000507712.1 |
KCNK16 |
potassium channel, subfamily K, member 16 |
| chr22_-_19974616 | 0.05 |
ENST00000344269.3 ENST00000401994.1 ENST00000406522.1 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr2_-_38604398 | 0.05 |
ENST00000443098.1 ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2 |
atlastin GTPase 2 |
| chr14_+_22670455 | 0.05 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr9_+_127539481 | 0.05 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr21_+_34775772 | 0.05 |
ENST00000405436.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr19_+_45582453 | 0.05 |
ENST00000591607.1 ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7 MARK4 |
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr20_-_23969416 | 0.05 |
ENST00000335694.4 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
| chr6_-_80247105 | 0.05 |
ENST00000369846.4 |
LCA5 |
Leber congenital amaurosis 5 |
| chr19_-_10444188 | 0.05 |
ENST00000293677.6 |
RAVER1 |
ribonucleoprotein, PTB-binding 1 |
| chr17_+_7788104 | 0.05 |
ENST00000380358.4 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr3_+_153839149 | 0.05 |
ENST00000465093.1 ENST00000465817.1 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_+_148928291 | 0.05 |
ENST00000420597.1 ENST00000452399.1 ENST00000294715.2 ENST00000539543.1 |
RP11-14N7.2 |
RP11-14N7.2 |
| chr17_-_74068707 | 0.05 |
ENST00000307877.2 |
SRP68 |
signal recognition particle 68kDa |
| chr20_+_48807351 | 0.05 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_-_5138099 | 0.05 |
ENST00000571800.1 ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
| chr1_-_115300579 | 0.05 |
ENST00000358528.4 ENST00000525132.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr1_-_24127256 | 0.05 |
ENST00000418277.1 |
GALE |
UDP-galactose-4-epimerase |
| chr2_+_228336849 | 0.05 |
ENST00000409979.2 ENST00000310078.8 |
AGFG1 |
ArfGAP with FG repeats 1 |
| chr1_-_201123586 | 0.05 |
ENST00000414605.2 ENST00000367334.5 ENST00000367332.1 |
TMEM9 |
transmembrane protein 9 |
| chrX_-_102319092 | 0.05 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr22_+_18834324 | 0.05 |
ENST00000342005.4 |
AC008132.13 |
Uncharacterized protein |
| chr1_+_150954493 | 0.05 |
ENST00000368947.4 |
ANXA9 |
annexin A9 |
| chrX_-_10557949 | 0.05 |
ENST00000380780.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr9_+_103189536 | 0.05 |
ENST00000374885.1 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
| chr15_-_90233866 | 0.05 |
ENST00000561257.1 |
PEX11A |
peroxisomal biogenesis factor 11 alpha |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 | 0.6 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.5 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 0.5 | GO:1904317 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.3 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.8 | GO:2001268 | positive regulation of keratinocyte migration(GO:0051549) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.2 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:0090677 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.2 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.2 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.5 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.0 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.4 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.2 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.5 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.1 | 0.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |