Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
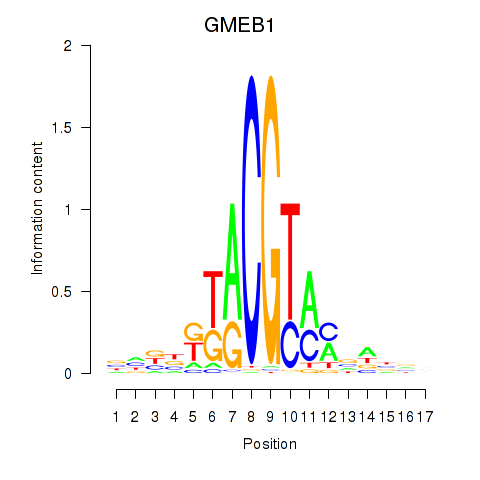
Results for GMEB1
Z-value: 0.51
Transcription factors associated with GMEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB1
|
ENSG00000162419.8 | GMEB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB1 | hg19_v2_chr1_+_28995258_28995322 | 0.65 | 8.1e-02 | Click! |
Activity profile of GMEB1 motif
Sorted Z-values of GMEB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_15850676 | 0.49 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr19_+_8478154 | 0.39 |
ENST00000381035.4 ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr1_+_163039143 | 0.39 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr5_+_156693091 | 0.38 |
ENST00000318218.6 ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr5_-_172756506 | 0.34 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr5_+_156693159 | 0.33 |
ENST00000347377.6 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_59010565 | 0.29 |
ENST00000594786.1 |
SLC27A5 |
solute carrier family 27 (fatty acid transporter), member 5 |
| chr16_-_4588762 | 0.29 |
ENST00000562334.1 ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr9_+_99690592 | 0.28 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr1_-_92371839 | 0.27 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr16_-_4588822 | 0.25 |
ENST00000564828.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr17_-_76870126 | 0.25 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr12_-_92539614 | 0.24 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr1_-_1293904 | 0.24 |
ENST00000309212.6 ENST00000342753.4 ENST00000445648.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr12_+_44152740 | 0.24 |
ENST00000440781.2 ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4 |
interleukin-1 receptor-associated kinase 4 |
| chr12_+_65672423 | 0.23 |
ENST00000355192.3 ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr12_-_124457257 | 0.22 |
ENST00000545891.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr22_+_35776828 | 0.22 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr16_-_67969888 | 0.21 |
ENST00000574576.2 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr7_+_139026057 | 0.21 |
ENST00000541515.3 |
LUC7L2 |
LUC7-like 2 (S. cerevisiae) |
| chr6_+_26204825 | 0.21 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr3_-_107941209 | 0.21 |
ENST00000492106.1 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr10_-_49812997 | 0.21 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr20_-_35402123 | 0.21 |
ENST00000373740.3 ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1 |
DSN1, MIS12 kinetochore complex component |
| chr5_+_40679584 | 0.20 |
ENST00000302472.3 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
| chr17_-_76870222 | 0.20 |
ENST00000585421.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr11_+_64879317 | 0.20 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr17_+_1665345 | 0.20 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr1_-_15850839 | 0.20 |
ENST00000348549.5 ENST00000546424.1 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr7_-_45960850 | 0.20 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr14_+_105941118 | 0.19 |
ENST00000550577.1 ENST00000538259.2 |
CRIP2 |
cysteine-rich protein 2 |
| chr9_-_73029540 | 0.19 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr4_-_2264015 | 0.19 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr21_-_38639601 | 0.19 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr15_-_91475706 | 0.19 |
ENST00000561036.1 |
HDDC3 |
HD domain containing 3 |
| chr17_-_46115122 | 0.18 |
ENST00000006101.4 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
| chr16_-_4588469 | 0.18 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr18_+_657733 | 0.18 |
ENST00000323250.5 ENST00000323224.7 |
TYMS |
thymidylate synthetase |
| chr19_+_57831829 | 0.18 |
ENST00000321545.4 |
ZNF543 |
zinc finger protein 543 |
| chr12_+_69202795 | 0.18 |
ENST00000539479.1 ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr11_-_114271139 | 0.18 |
ENST00000325636.4 |
C11orf71 |
chromosome 11 open reading frame 71 |
| chr19_-_17414179 | 0.18 |
ENST00000594194.1 ENST00000247706.3 |
ABHD8 |
abhydrolase domain containing 8 |
| chr10_-_79397391 | 0.18 |
ENST00000286628.8 ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_221054584 | 0.17 |
ENST00000549319.1 |
HLX |
H2.0-like homeobox |
| chr21_-_45078019 | 0.17 |
ENST00000542962.1 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr15_+_33010175 | 0.17 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_-_92928557 | 0.17 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr5_+_178450753 | 0.17 |
ENST00000444149.2 ENST00000519896.1 ENST00000522442.1 |
ZNF879 |
zinc finger protein 879 |
| chr20_+_49411543 | 0.17 |
ENST00000609336.1 ENST00000445038.1 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr3_+_54156570 | 0.17 |
ENST00000415676.2 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr1_-_95392635 | 0.17 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chr20_+_42219559 | 0.17 |
ENST00000373030.3 ENST00000373039.4 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr19_+_18668572 | 0.17 |
ENST00000540691.1 ENST00000539106.1 ENST00000222307.4 |
KXD1 |
KxDL motif containing 1 |
| chrX_+_51928002 | 0.16 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr6_-_26189304 | 0.16 |
ENST00000340756.2 |
HIST1H4D |
histone cluster 1, H4d |
| chr17_+_17206635 | 0.16 |
ENST00000389022.4 |
NT5M |
5',3'-nucleotidase, mitochondrial |
| chr7_-_50860565 | 0.16 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr10_-_13342097 | 0.15 |
ENST00000263038.4 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
| chr2_-_9563469 | 0.15 |
ENST00000484735.1 ENST00000456913.2 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr7_-_56119156 | 0.15 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr12_+_7052974 | 0.15 |
ENST00000544681.1 ENST00000537087.1 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr14_-_106406090 | 0.15 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chr1_+_161284047 | 0.15 |
ENST00000367975.2 ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC |
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr7_+_99746514 | 0.15 |
ENST00000341942.5 ENST00000441173.1 |
LAMTOR4 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr6_+_32821924 | 0.15 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_-_100025238 | 0.15 |
ENST00000521696.1 |
RP11-410L14.2 |
RP11-410L14.2 |
| chr11_+_65601112 | 0.15 |
ENST00000308342.6 |
SNX32 |
sorting nexin 32 |
| chr5_-_107717058 | 0.14 |
ENST00000359660.5 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr2_+_56411131 | 0.14 |
ENST00000407595.2 |
CCDC85A |
coiled-coil domain containing 85A |
| chr12_+_7053172 | 0.14 |
ENST00000229281.5 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr5_+_130599701 | 0.14 |
ENST00000395246.1 |
CDC42SE2 |
CDC42 small effector 2 |
| chr16_+_67204400 | 0.14 |
ENST00000563439.1 ENST00000432069.2 ENST00000564992.1 ENST00000564053.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr16_+_15596123 | 0.14 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr20_+_43160409 | 0.14 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_+_145516560 | 0.14 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr6_-_52859046 | 0.14 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr18_-_70534745 | 0.14 |
ENST00000583169.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr20_+_43160458 | 0.14 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr8_-_99129338 | 0.14 |
ENST00000520507.1 |
HRSP12 |
heat-responsive protein 12 |
| chr1_-_154909329 | 0.14 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr16_+_3550924 | 0.14 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr4_-_184580353 | 0.14 |
ENST00000326397.5 |
RWDD4 |
RWD domain containing 4 |
| chr15_+_57998821 | 0.14 |
ENST00000299638.3 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr7_+_1748798 | 0.14 |
ENST00000561626.1 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr6_+_116892530 | 0.14 |
ENST00000466444.2 ENST00000368590.5 ENST00000392526.1 |
RWDD1 |
RWD domain containing 1 |
| chr1_+_61547894 | 0.13 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr2_+_201981527 | 0.13 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr19_-_58951496 | 0.13 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr3_-_107941230 | 0.13 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_+_149871135 | 0.13 |
ENST00000369152.5 |
BOLA1 |
bolA family member 1 |
| chr1_+_47799542 | 0.13 |
ENST00000471289.2 ENST00000450808.2 |
CMPK1 |
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr14_+_74035763 | 0.13 |
ENST00000238651.5 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr22_-_19435209 | 0.13 |
ENST00000546308.1 ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA C22orf39 |
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr15_+_41056255 | 0.13 |
ENST00000561160.1 ENST00000559445.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr22_-_36877371 | 0.13 |
ENST00000403313.1 |
TXN2 |
thioredoxin 2 |
| chr1_-_28969517 | 0.13 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr12_+_69202975 | 0.13 |
ENST00000544561.1 ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr19_+_1104048 | 0.13 |
ENST00000593032.1 ENST00000588919.1 |
GPX4 |
glutathione peroxidase 4 |
| chr5_-_171433819 | 0.13 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr15_+_41056218 | 0.13 |
ENST00000260447.4 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr16_-_3073933 | 0.13 |
ENST00000574151.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr17_-_9479128 | 0.13 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr21_+_45553535 | 0.13 |
ENST00000348499.5 ENST00000389690.3 ENST00000449622.1 |
C21orf33 |
chromosome 21 open reading frame 33 |
| chr1_-_1310530 | 0.13 |
ENST00000338370.3 ENST00000321751.5 ENST00000378853.3 |
AURKAIP1 |
aurora kinase A interacting protein 1 |
| chr6_-_31107127 | 0.13 |
ENST00000259845.4 |
PSORS1C2 |
psoriasis susceptibility 1 candidate 2 |
| chr1_+_228327943 | 0.13 |
ENST00000366726.1 ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1 |
guanylate kinase 1 |
| chr5_-_139944196 | 0.13 |
ENST00000357560.4 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr16_+_68573116 | 0.13 |
ENST00000570495.1 ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90 |
ZFP90 zinc finger protein |
| chrX_-_134478012 | 0.13 |
ENST00000370766.3 |
ZNF75D |
zinc finger protein 75D |
| chr5_+_178986693 | 0.13 |
ENST00000437570.2 ENST00000393438.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr2_-_20101701 | 0.13 |
ENST00000402414.1 ENST00000333610.3 |
TTC32 |
tetratricopeptide repeat domain 32 |
| chr1_+_197170592 | 0.13 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chrX_+_12993336 | 0.13 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr11_-_105948040 | 0.13 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr12_+_7053228 | 0.12 |
ENST00000540506.2 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr19_+_42364460 | 0.12 |
ENST00000593863.1 |
RPS19 |
ribosomal protein S19 |
| chr3_+_54156664 | 0.12 |
ENST00000474759.1 ENST00000288197.5 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr14_+_50779071 | 0.12 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr11_+_124609742 | 0.12 |
ENST00000284292.6 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr8_+_91013676 | 0.12 |
ENST00000519410.1 ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr19_+_1103936 | 0.12 |
ENST00000354171.8 ENST00000589115.1 |
GPX4 |
glutathione peroxidase 4 |
| chr12_+_50505762 | 0.12 |
ENST00000550487.1 ENST00000317943.2 |
COX14 |
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr12_+_50505963 | 0.12 |
ENST00000550654.1 ENST00000548985.1 |
COX14 |
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr5_+_130599735 | 0.12 |
ENST00000503291.1 ENST00000360515.3 ENST00000505065.1 |
CDC42SE2 |
CDC42 small effector 2 |
| chr4_+_108910870 | 0.12 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr2_+_106682135 | 0.12 |
ENST00000437659.1 |
C2orf40 |
chromosome 2 open reading frame 40 |
| chr5_+_154238042 | 0.12 |
ENST00000519211.1 ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr14_-_53258314 | 0.12 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr11_-_105948129 | 0.12 |
ENST00000526793.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr15_+_55700741 | 0.12 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr4_+_154074217 | 0.12 |
ENST00000437508.2 |
TRIM2 |
tripartite motif containing 2 |
| chr11_+_133938820 | 0.12 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr16_+_56716336 | 0.12 |
ENST00000394485.4 ENST00000562939.1 |
MT1X |
metallothionein 1X |
| chr9_+_34329493 | 0.12 |
ENST00000379158.2 ENST00000346365.4 ENST00000379155.5 |
NUDT2 |
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr8_-_99129384 | 0.12 |
ENST00000521560.1 ENST00000254878.3 |
HRSP12 |
heat-responsive protein 12 |
| chr1_-_36916066 | 0.12 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr22_-_19166343 | 0.12 |
ENST00000215882.5 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr8_+_81397876 | 0.11 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr5_-_178054014 | 0.11 |
ENST00000520957.1 |
CLK4 |
CDC-like kinase 4 |
| chrX_+_12993202 | 0.11 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr20_+_44563267 | 0.11 |
ENST00000372409.3 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
| chr17_-_38083843 | 0.11 |
ENST00000304046.2 ENST00000579695.1 |
ORMDL3 |
ORM1-like 3 (S. cerevisiae) |
| chr16_-_15149828 | 0.11 |
ENST00000566419.1 ENST00000568320.1 |
NTAN1 |
N-terminal asparagine amidase |
| chr10_-_90751038 | 0.11 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr19_+_18668672 | 0.11 |
ENST00000601630.1 ENST00000599000.1 ENST00000595073.1 ENST00000596785.1 |
KXD1 |
KxDL motif containing 1 |
| chr19_+_44037546 | 0.11 |
ENST00000601282.1 |
ZNF575 |
zinc finger protein 575 |
| chr17_+_36908984 | 0.11 |
ENST00000225426.4 ENST00000579088.1 |
PSMB3 |
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr1_-_19229248 | 0.11 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr1_+_210502238 | 0.11 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr3_-_197676740 | 0.11 |
ENST00000452735.1 ENST00000453254.1 ENST00000455191.1 |
IQCG |
IQ motif containing G |
| chr7_-_100965011 | 0.11 |
ENST00000498704.2 ENST00000517481.1 ENST00000437644.2 ENST00000315322.4 |
RABL5 |
RAB, member RAS oncogene family-like 5 |
| chr11_-_111749878 | 0.11 |
ENST00000260257.4 |
FDXACB1 |
ferredoxin-fold anticodon binding domain containing 1 |
| chr15_+_57998923 | 0.11 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr11_+_20044600 | 0.11 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr22_-_41864662 | 0.11 |
ENST00000216252.3 |
PHF5A |
PHD finger protein 5A |
| chr16_-_88729473 | 0.11 |
ENST00000301012.3 ENST00000569177.1 |
MVD |
mevalonate (diphospho) decarboxylase |
| chr2_+_71295733 | 0.11 |
ENST00000443938.2 ENST00000244204.6 |
NAGK |
N-acetylglucosamine kinase |
| chr14_-_75079026 | 0.11 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
| chr16_+_3074002 | 0.11 |
ENST00000326266.8 ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6 |
THO complex 6 homolog (Drosophila) |
| chr22_-_19435755 | 0.11 |
ENST00000542103.1 ENST00000399562.4 |
C22orf39 |
chromosome 22 open reading frame 39 |
| chr22_-_50699701 | 0.11 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr10_+_135192695 | 0.11 |
ENST00000368539.4 ENST00000278060.5 ENST00000357296.3 |
PAOX |
polyamine oxidase (exo-N4-amino) |
| chr13_-_28024681 | 0.11 |
ENST00000381116.1 ENST00000381120.3 ENST00000431572.2 |
MTIF3 |
mitochondrial translational initiation factor 3 |
| chr1_-_155881156 | 0.11 |
ENST00000539040.1 ENST00000368323.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr11_+_69455855 | 0.11 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr5_-_10761206 | 0.11 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr17_+_38296576 | 0.11 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr18_+_657578 | 0.11 |
ENST00000323274.10 |
TYMS |
thymidylate synthetase |
| chr6_+_157802165 | 0.11 |
ENST00000414563.2 ENST00000359775.5 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
| chr8_+_13424352 | 0.11 |
ENST00000297324.4 |
C8orf48 |
chromosome 8 open reading frame 48 |
| chr16_-_3074231 | 0.10 |
ENST00000572355.1 ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr22_-_30162924 | 0.10 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr7_-_100888313 | 0.10 |
ENST00000442303.1 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr3_+_134204881 | 0.10 |
ENST00000511574.1 ENST00000337090.3 ENST00000383229.3 |
CEP63 |
centrosomal protein 63kDa |
| chr3_+_111697843 | 0.10 |
ENST00000534857.1 ENST00000273359.3 ENST00000494817.1 |
ABHD10 |
abhydrolase domain containing 10 |
| chr17_+_1665253 | 0.10 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_893200 | 0.10 |
ENST00000269814.4 ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16 |
mediator complex subunit 16 |
| chr3_-_182833863 | 0.10 |
ENST00000492597.1 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr1_+_201617264 | 0.10 |
ENST00000367296.4 |
NAV1 |
neuron navigator 1 |
| chr5_-_93447333 | 0.10 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr13_-_114898016 | 0.10 |
ENST00000542651.1 ENST00000334062.7 |
RASA3 |
RAS p21 protein activator 3 |
| chr1_+_25944341 | 0.10 |
ENST00000263979.3 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr19_+_1104415 | 0.10 |
ENST00000585362.2 |
GPX4 |
glutathione peroxidase 4 |
| chr11_+_95523823 | 0.10 |
ENST00000538658.1 |
CEP57 |
centrosomal protein 57kDa |
| chr22_-_38902300 | 0.10 |
ENST00000403230.1 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
| chr6_+_28092338 | 0.10 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr19_+_33463210 | 0.10 |
ENST00000590281.1 |
C19orf40 |
chromosome 19 open reading frame 40 |
| chr7_+_73703728 | 0.10 |
ENST00000361545.5 ENST00000223398.6 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
| chr1_+_228327923 | 0.10 |
ENST00000391865.3 |
GUK1 |
guanylate kinase 1 |
| chr8_-_64080945 | 0.10 |
ENST00000603538.1 |
YTHDF3-AS1 |
YTHDF3 antisense RNA 1 (head to head) |
| chr8_+_26240414 | 0.10 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_-_31769656 | 0.10 |
ENST00000446633.2 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr10_-_127511790 | 0.10 |
ENST00000368797.4 ENST00000420761.1 |
UROS |
uroporphyrinogen III synthase |
| chr1_-_19229014 | 0.10 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr11_-_67276100 | 0.10 |
ENST00000301488.3 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr13_+_111972980 | 0.10 |
ENST00000283547.1 |
TEX29 |
testis expressed 29 |
| chr12_+_107349497 | 0.10 |
ENST00000548125.1 ENST00000280756.4 |
C12orf23 |
chromosome 12 open reading frame 23 |
| chr7_-_27702455 | 0.10 |
ENST00000265395.2 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
| chr3_-_46506358 | 0.10 |
ENST00000417439.1 ENST00000431944.1 |
LTF |
lactotransferrin |
| chr12_-_6772303 | 0.10 |
ENST00000396807.4 ENST00000446105.2 ENST00000341550.4 |
ING4 |
inhibitor of growth family, member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.3 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.2 | GO:0032764 | smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.3 | GO:0019860 | transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.2 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.2 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.1 | GO:1900157 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) regulation of bone mineralization involved in bone maturation(GO:1900157) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0060661 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.2 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.0 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0071611 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0009450 | acetate metabolic process(GO:0006083) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.0 | GO:0019322 | xylulose metabolic process(GO:0005997) pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of endosome membrane(GO:0010009) cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.0 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0031403 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |