Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for GMEB2
Z-value: 0.11
Transcription factors associated with GMEB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB2
|
ENSG00000101216.6 | GMEB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | hg19_v2_chr20_-_62258394_62258464 | -0.59 | 1.3e-01 | Click! |
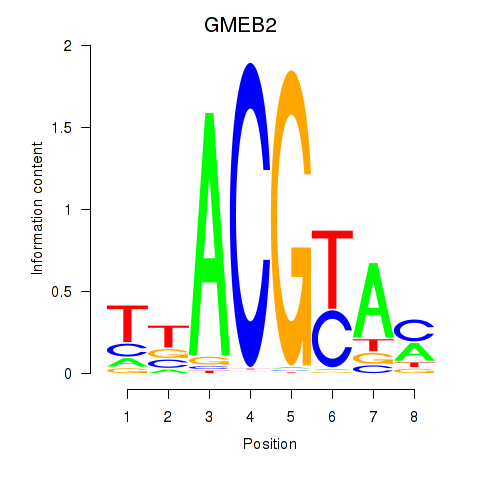
Activity profile of GMEB2 motif
Sorted Z-values of GMEB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_65667884 | 0.33 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr7_-_19748640 | 0.30 |
ENST00000222567.5 |
TWISTNB |
TWIST neighbor |
| chr11_-_65667997 | 0.29 |
ENST00000312562.2 ENST00000534222.1 |
FOSL1 |
FOS-like antigen 1 |
| chr7_-_73153122 | 0.22 |
ENST00000458339.1 |
ABHD11 |
abhydrolase domain containing 11 |
| chr16_+_3074002 | 0.22 |
ENST00000326266.8 ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6 |
THO complex 6 homolog (Drosophila) |
| chr6_+_25992884 | 0.21 |
ENST00000606723.2 |
U91328.19 |
U91328.19 |
| chr7_-_45151272 | 0.21 |
ENST00000461363.1 ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4 |
transforming growth factor beta regulator 4 |
| chr7_-_73153178 | 0.20 |
ENST00000437775.2 ENST00000222800.3 |
ABHD11 |
abhydrolase domain containing 11 |
| chr8_-_134309823 | 0.19 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr8_-_125384927 | 0.19 |
ENST00000297632.6 |
TMEM65 |
transmembrane protein 65 |
| chr15_+_90118685 | 0.19 |
ENST00000268138.7 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
| chr6_-_27840099 | 0.19 |
ENST00000328488.2 |
HIST1H3I |
histone cluster 1, H3i |
| chr19_+_4402659 | 0.19 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_101869262 | 0.18 |
ENST00000289382.3 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
| chr5_-_74162605 | 0.18 |
ENST00000389156.4 ENST00000510496.1 ENST00000380515.3 |
FAM169A |
family with sequence similarity 169, member A |
| chr7_-_73153161 | 0.18 |
ENST00000395147.4 |
ABHD11 |
abhydrolase domain containing 11 |
| chr15_+_90118723 | 0.18 |
ENST00000560985.1 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
| chr19_+_10765003 | 0.18 |
ENST00000407004.3 ENST00000589998.1 ENST00000589600.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr1_-_52870059 | 0.18 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr17_+_66511540 | 0.18 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr12_+_122064673 | 0.17 |
ENST00000537188.1 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr1_+_234509186 | 0.17 |
ENST00000366615.4 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr14_-_53258314 | 0.17 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr5_-_176057518 | 0.17 |
ENST00000393693.2 |
SNCB |
synuclein, beta |
| chr2_-_46769694 | 0.17 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr7_-_56119156 | 0.16 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr17_+_38296576 | 0.16 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr1_+_65613340 | 0.16 |
ENST00000546702.1 |
AK4 |
adenylate kinase 4 |
| chr8_+_38243951 | 0.16 |
ENST00000297720.5 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_+_4791722 | 0.16 |
ENST00000269856.3 |
FEM1A |
fem-1 homolog a (C. elegans) |
| chr6_+_26204825 | 0.15 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr4_+_113152978 | 0.15 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr12_+_118814185 | 0.15 |
ENST00000543473.1 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr1_-_52870104 | 0.15 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr12_+_56360605 | 0.15 |
ENST00000553376.1 ENST00000440311.2 ENST00000354056.4 |
CDK2 |
cyclin-dependent kinase 2 |
| chr11_+_60609537 | 0.15 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr20_+_361890 | 0.15 |
ENST00000449710.1 ENST00000422053.2 |
TRIB3 |
tribbles pseudokinase 3 |
| chr13_+_48611665 | 0.14 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr15_-_72490114 | 0.14 |
ENST00000309731.7 |
GRAMD2 |
GRAM domain containing 2 |
| chr12_+_22778291 | 0.14 |
ENST00000545979.1 |
ETNK1 |
ethanolamine kinase 1 |
| chr5_-_176057365 | 0.14 |
ENST00000310112.3 |
SNCB |
synuclein, beta |
| chr14_+_53196872 | 0.14 |
ENST00000442123.2 ENST00000354586.4 |
STYX |
serine/threonine/tyrosine interacting protein |
| chr10_+_60145155 | 0.14 |
ENST00000373895.3 |
TFAM |
transcription factor A, mitochondrial |
| chr18_-_19748331 | 0.14 |
ENST00000584201.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr19_-_663277 | 0.14 |
ENST00000292363.5 |
RNF126 |
ring finger protein 126 |
| chr18_-_19748379 | 0.14 |
ENST00000579431.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr1_-_31769656 | 0.14 |
ENST00000446633.2 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr19_+_9296279 | 0.13 |
ENST00000344248.2 |
OR7D2 |
olfactory receptor, family 7, subfamily D, member 2 |
| chr11_-_111749767 | 0.13 |
ENST00000542429.1 |
FDXACB1 |
ferredoxin-fold anticodon binding domain containing 1 |
| chr17_-_74528128 | 0.13 |
ENST00000590175.1 |
CYGB |
cytoglobin |
| chr12_+_53662110 | 0.13 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr11_-_111749878 | 0.13 |
ENST00000260257.4 |
FDXACB1 |
ferredoxin-fold anticodon binding domain containing 1 |
| chr18_+_74240610 | 0.13 |
ENST00000578092.1 ENST00000578613.1 ENST00000583578.1 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
| chr8_-_124408652 | 0.13 |
ENST00000287394.5 |
ATAD2 |
ATPase family, AAA domain containing 2 |
| chr19_+_49458107 | 0.13 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr12_+_7079944 | 0.13 |
ENST00000261406.6 |
EMG1 |
EMG1 N1-specific pseudouridine methyltransferase |
| chr2_+_227700652 | 0.13 |
ENST00000341329.3 ENST00000392062.2 ENST00000437454.1 ENST00000443477.1 ENST00000423616.1 ENST00000448992.1 |
RHBDD1 |
rhomboid domain containing 1 |
| chr10_-_27529486 | 0.13 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr10_+_13203543 | 0.13 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chrX_+_133507327 | 0.12 |
ENST00000332070.3 ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6 |
PHD finger protein 6 |
| chrX_-_48755030 | 0.12 |
ENST00000490755.2 ENST00000465150.2 ENST00000495490.2 |
TIMM17B |
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr8_+_38243967 | 0.12 |
ENST00000524874.1 ENST00000379957.4 ENST00000523983.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_75311019 | 0.12 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr2_-_100939195 | 0.12 |
ENST00000393437.3 |
LONRF2 |
LON peptidase N-terminal domain and ring finger 2 |
| chr1_-_161087802 | 0.12 |
ENST00000368010.3 |
PFDN2 |
prefoldin subunit 2 |
| chr9_+_138235095 | 0.12 |
ENST00000320778.2 |
C9orf62 |
chromosome 9 open reading frame 62 |
| chr18_+_77439775 | 0.12 |
ENST00000299543.7 ENST00000075430.7 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr8_-_134309335 | 0.12 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr1_-_31769595 | 0.12 |
ENST00000263694.4 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr22_-_42017021 | 0.12 |
ENST00000263256.6 |
DESI1 |
desumoylating isopeptidase 1 |
| chr4_+_39460689 | 0.12 |
ENST00000381846.1 |
LIAS |
lipoic acid synthetase |
| chr5_+_94890778 | 0.12 |
ENST00000380009.4 |
ARSK |
arylsulfatase family, member K |
| chrX_+_48660287 | 0.12 |
ENST00000444343.2 ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6 |
histone deacetylase 6 |
| chr4_+_113152881 | 0.11 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr15_-_73075964 | 0.11 |
ENST00000563907.1 |
ADPGK |
ADP-dependent glucokinase |
| chrX_+_133507389 | 0.11 |
ENST00000370800.4 |
PHF6 |
PHD finger protein 6 |
| chr8_-_80942467 | 0.11 |
ENST00000518271.1 ENST00000276585.4 ENST00000521605.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr4_+_39460659 | 0.11 |
ENST00000513731.1 |
LIAS |
lipoic acid synthetase |
| chr4_+_39460620 | 0.11 |
ENST00000340169.2 ENST00000261434.3 |
LIAS |
lipoic acid synthetase |
| chr1_-_109203997 | 0.11 |
ENST00000370032.5 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr8_+_118533049 | 0.11 |
ENST00000522839.1 |
MED30 |
mediator complex subunit 30 |
| chr1_-_200589859 | 0.11 |
ENST00000367350.4 |
KIF14 |
kinesin family member 14 |
| chr19_+_17830051 | 0.11 |
ENST00000594625.1 ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S |
microtubule-associated protein 1S |
| chr10_-_135379132 | 0.11 |
ENST00000343131.5 |
SYCE1 |
synaptonemal complex central element protein 1 |
| chrX_+_133507283 | 0.11 |
ENST00000370803.3 |
PHF6 |
PHD finger protein 6 |
| chr10_+_35415851 | 0.11 |
ENST00000374726.3 |
CREM |
cAMP responsive element modulator |
| chr3_+_50654550 | 0.11 |
ENST00000430409.1 ENST00000357955.2 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
| chr15_+_40453204 | 0.11 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr5_+_82373317 | 0.11 |
ENST00000282268.3 ENST00000338635.6 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr10_+_60144782 | 0.11 |
ENST00000487519.1 |
TFAM |
transcription factor A, mitochondrial |
| chr4_+_75310851 | 0.11 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr10_+_70939983 | 0.11 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr2_+_118572226 | 0.10 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr2_+_54683419 | 0.10 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr11_+_33279850 | 0.10 |
ENST00000531504.1 ENST00000456517.1 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr1_-_713985 | 0.10 |
ENST00000428504.1 |
RP11-206L10.2 |
RP11-206L10.2 |
| chr1_+_22333943 | 0.10 |
ENST00000400271.2 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chr16_+_3115298 | 0.10 |
ENST00000325568.5 ENST00000534507.1 |
IL32 |
interleukin 32 |
| chr21_-_34914394 | 0.10 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr6_-_136571400 | 0.10 |
ENST00000418509.2 ENST00000420702.1 ENST00000451457.2 |
MTFR2 |
mitochondrial fission regulator 2 |
| chr16_-_75590114 | 0.10 |
ENST00000568377.1 ENST00000565067.1 ENST00000258173.6 |
TMEM231 |
transmembrane protein 231 |
| chr4_+_178230985 | 0.10 |
ENST00000264596.3 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
| chr3_-_47823298 | 0.10 |
ENST00000254480.5 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr16_+_3115611 | 0.10 |
ENST00000530890.1 ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32 |
interleukin 32 |
| chr3_-_185542817 | 0.10 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr16_-_46655538 | 0.10 |
ENST00000303383.3 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
| chr4_-_113558014 | 0.10 |
ENST00000503172.1 ENST00000505019.1 ENST00000309071.5 |
C4orf21 |
chromosome 4 open reading frame 21 |
| chr3_-_9005118 | 0.10 |
ENST00000264926.2 |
RAD18 |
RAD18 homolog (S. cerevisiae) |
| chr16_+_31539197 | 0.10 |
ENST00000564707.1 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr15_-_35280426 | 0.10 |
ENST00000559564.1 ENST00000356321.4 |
ZNF770 |
zinc finger protein 770 |
| chr15_+_81293254 | 0.10 |
ENST00000267984.2 |
MESDC1 |
mesoderm development candidate 1 |
| chr19_+_52693259 | 0.10 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr15_+_66797627 | 0.10 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr12_+_102514019 | 0.09 |
ENST00000537257.1 ENST00000358383.5 ENST00000392911.2 |
PARPBP |
PARP1 binding protein |
| chr12_+_118814344 | 0.09 |
ENST00000397564.2 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr2_+_88991162 | 0.09 |
ENST00000283646.4 |
RPIA |
ribose 5-phosphate isomerase A |
| chr22_+_21271714 | 0.09 |
ENST00000354336.3 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr7_+_5632436 | 0.09 |
ENST00000340250.6 ENST00000382361.3 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chrX_+_21958674 | 0.09 |
ENST00000404933.2 |
SMS |
spermine synthase |
| chr2_+_178077477 | 0.09 |
ENST00000411529.2 ENST00000435711.1 |
HNRNPA3 |
heterogeneous nuclear ribonucleoprotein A3 |
| chr12_+_53662073 | 0.09 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr12_+_102513950 | 0.09 |
ENST00000378128.3 ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP |
PARP1 binding protein |
| chr3_+_124449213 | 0.09 |
ENST00000232607.2 ENST00000536109.1 ENST00000538242.1 ENST00000413078.2 |
UMPS |
uridine monophosphate synthetase |
| chr14_-_21737610 | 0.09 |
ENST00000320084.7 ENST00000449098.1 ENST00000336053.6 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr5_-_16916624 | 0.09 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr1_+_23345930 | 0.09 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr7_-_97501706 | 0.09 |
ENST00000455086.1 ENST00000453600.1 |
ASNS |
asparagine synthetase (glutamine-hydrolyzing) |
| chr14_+_62229075 | 0.09 |
ENST00000216294.4 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr14_-_64108537 | 0.09 |
ENST00000554717.1 ENST00000394942.2 |
WDR89 |
WD repeat domain 89 |
| chr3_-_14166316 | 0.09 |
ENST00000396914.3 ENST00000295767.5 |
CHCHD4 |
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr14_+_102786063 | 0.09 |
ENST00000442396.2 ENST00000262236.5 |
ZNF839 |
zinc finger protein 839 |
| chr1_+_23345943 | 0.09 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr16_+_31044413 | 0.09 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr1_+_153606513 | 0.09 |
ENST00000368694.3 |
CHTOP |
chromatin target of PRMT1 |
| chr18_-_2571437 | 0.09 |
ENST00000574676.1 ENST00000574538.1 ENST00000319888.6 |
METTL4 |
methyltransferase like 4 |
| chr16_+_19079215 | 0.09 |
ENST00000544894.2 ENST00000561858.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr12_+_94071129 | 0.09 |
ENST00000552983.1 ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr2_+_234160217 | 0.09 |
ENST00000392017.4 ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr5_-_122759032 | 0.09 |
ENST00000510582.3 ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120 |
centrosomal protein 120kDa |
| chr12_+_94071341 | 0.09 |
ENST00000542893.2 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr15_+_66797455 | 0.09 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr22_-_38902300 | 0.08 |
ENST00000403230.1 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
| chr2_-_44588893 | 0.08 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr3_-_196669298 | 0.08 |
ENST00000411704.1 ENST00000452404.2 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
| chr16_+_31044812 | 0.08 |
ENST00000313843.3 |
STX4 |
syntaxin 4 |
| chr8_-_80942061 | 0.08 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr6_+_30029008 | 0.08 |
ENST00000332435.5 ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1 |
zinc ribbon domain containing 1 |
| chr19_-_36399149 | 0.08 |
ENST00000585901.2 ENST00000544690.2 ENST00000424586.3 ENST00000262629.4 ENST00000589517.1 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
| chr8_-_80942139 | 0.08 |
ENST00000521434.1 ENST00000519120.1 ENST00000520946.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr20_+_60697480 | 0.08 |
ENST00000370915.1 ENST00000253001.4 ENST00000400318.2 ENST00000279068.6 ENST00000279069.7 |
LSM14B |
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr17_+_7462031 | 0.08 |
ENST00000380535.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr14_-_20020272 | 0.08 |
ENST00000551509.1 |
POTEM |
POTE ankyrin domain family, member M |
| chr19_+_18942720 | 0.08 |
ENST00000262803.5 |
UPF1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr19_+_10764937 | 0.08 |
ENST00000449870.1 ENST00000318511.3 ENST00000420083.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr1_+_173837214 | 0.08 |
ENST00000367704.1 |
ZBTB37 |
zinc finger and BTB domain containing 37 |
| chr16_+_19078960 | 0.08 |
ENST00000568985.1 ENST00000566110.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr5_-_32313019 | 0.08 |
ENST00000280285.5 ENST00000264934.5 |
MTMR12 |
myotubularin related protein 12 |
| chr20_+_56136136 | 0.08 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chrX_+_41193407 | 0.08 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr17_-_40306934 | 0.08 |
ENST00000592574.1 ENST00000550406.1 ENST00000547517.1 ENST00000393860.3 ENST00000346213.4 |
CTD-2132N18.3 RAB5C |
Uncharacterized protein RAB5C, member RAS oncogene family |
| chr2_+_232572361 | 0.08 |
ENST00000409321.1 |
PTMA |
prothymosin, alpha |
| chr12_+_50017184 | 0.08 |
ENST00000548825.2 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr5_+_94890840 | 0.08 |
ENST00000504763.1 |
ARSK |
arylsulfatase family, member K |
| chr5_-_176056974 | 0.08 |
ENST00000510387.1 ENST00000506696.1 |
SNCB |
synuclein, beta |
| chr12_+_56360550 | 0.08 |
ENST00000266970.4 |
CDK2 |
cyclin-dependent kinase 2 |
| chr8_+_48873453 | 0.08 |
ENST00000523944.1 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr15_-_52861157 | 0.08 |
ENST00000564163.1 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
| chr6_-_55443958 | 0.08 |
ENST00000370850.2 |
HMGCLL1 |
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr8_+_48873479 | 0.08 |
ENST00000262105.2 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr11_+_43333513 | 0.08 |
ENST00000534695.1 ENST00000455725.2 ENST00000531273.1 ENST00000420461.2 ENST00000378852.3 ENST00000534600.1 |
API5 |
apoptosis inhibitor 5 |
| chr5_-_122758994 | 0.08 |
ENST00000306467.5 ENST00000515110.1 |
CEP120 |
centrosomal protein 120kDa |
| chr3_-_132378897 | 0.08 |
ENST00000545291.1 |
ACAD11 |
acyl-CoA dehydrogenase family, member 11 |
| chr12_-_82752159 | 0.08 |
ENST00000552377.1 |
CCDC59 |
coiled-coil domain containing 59 |
| chr15_+_77712993 | 0.08 |
ENST00000336216.4 ENST00000381714.3 ENST00000558651.1 |
HMG20A |
high mobility group 20A |
| chr1_+_173837488 | 0.08 |
ENST00000427304.1 ENST00000432989.1 ENST00000367702.1 |
ZBTB37 |
zinc finger and BTB domain containing 37 |
| chr6_-_135375921 | 0.08 |
ENST00000367820.2 ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L |
HBS1-like (S. cerevisiae) |
| chr17_+_7465216 | 0.07 |
ENST00000321337.7 |
SENP3 |
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr10_-_27529716 | 0.07 |
ENST00000375897.3 ENST00000396271.3 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr14_-_21737551 | 0.07 |
ENST00000554891.1 ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr9_-_77643307 | 0.07 |
ENST00000376834.3 ENST00000376830.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr2_+_234160340 | 0.07 |
ENST00000417017.1 ENST00000392020.4 ENST00000392018.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr19_-_41903161 | 0.07 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr9_-_139137648 | 0.07 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr16_+_19079311 | 0.07 |
ENST00000569127.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr11_+_65337901 | 0.07 |
ENST00000309328.3 ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1 |
Sjogren syndrome/scleroderma autoantigen 1 |
| chr14_-_95786200 | 0.07 |
ENST00000298912.4 |
CLMN |
calmin (calponin-like, transmembrane) |
| chr7_-_73668692 | 0.07 |
ENST00000352131.3 ENST00000055077.3 |
RFC2 |
replication factor C (activator 1) 2, 40kDa |
| chr2_+_234600253 | 0.07 |
ENST00000373424.1 ENST00000441351.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_65215604 | 0.07 |
ENST00000531327.1 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_162887556 | 0.07 |
ENST00000393915.4 ENST00000432118.2 ENST00000358715.3 |
HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
| chrX_+_41192595 | 0.07 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr16_-_20681177 | 0.07 |
ENST00000524149.1 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chrY_+_15016725 | 0.07 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr19_-_6767516 | 0.07 |
ENST00000245908.6 |
SH2D3A |
SH2 domain containing 3A |
| chr16_-_103572 | 0.07 |
ENST00000293860.5 |
POLR3K |
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr5_-_32313082 | 0.07 |
ENST00000382142.3 |
MTMR12 |
myotubularin related protein 12 |
| chr10_+_91461337 | 0.07 |
ENST00000260753.4 ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B |
kinesin family member 20B |
| chr12_+_6603253 | 0.07 |
ENST00000382457.4 ENST00000545962.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr3_-_195808980 | 0.07 |
ENST00000360110.4 |
TFRC |
transferrin receptor |
| chrX_-_77150911 | 0.07 |
ENST00000373336.3 |
MAGT1 |
magnesium transporter 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:1902339 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0032986 | protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0060600 | dichotomous subdivision of an epithelial terminal unit(GO:0060600) |
| 0.0 | 0.0 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.0 | GO:0072429 | meiotic DNA double-strand break processing(GO:0000706) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097124 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.1 | 0.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |