Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for HIF1A
Z-value: 0.25
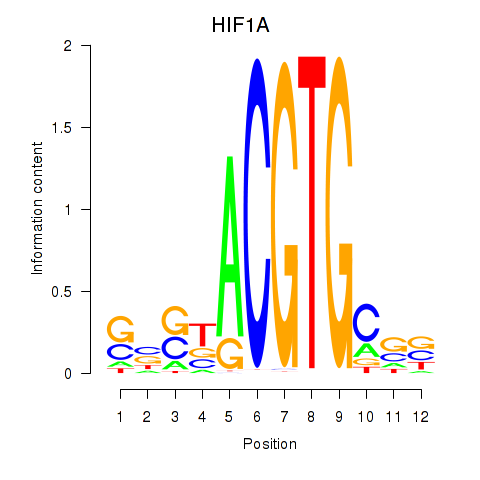
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | HIF1A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg19_v2_chr14_+_62162258_62162269 | 0.75 | 3.4e-02 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_93392811 | 0.18 |
ENST00000238994.5 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
| chr1_-_92351769 | 0.17 |
ENST00000212355.4 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr8_+_17354617 | 0.15 |
ENST00000470360.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr21_+_26934165 | 0.14 |
ENST00000456917.1 |
MIR155HG |
MIR155 host gene (non-protein coding) |
| chr10_-_49732281 | 0.14 |
ENST00000374170.1 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr8_+_17354587 | 0.13 |
ENST00000494857.1 ENST00000522656.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr3_-_71834318 | 0.12 |
ENST00000353065.3 |
PROK2 |
prokineticin 2 |
| chr8_-_95908902 | 0.11 |
ENST00000520509.1 |
CCNE2 |
cyclin E2 |
| chr4_-_39529049 | 0.11 |
ENST00000501493.2 ENST00000509391.1 ENST00000507089.1 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr4_-_39529180 | 0.11 |
ENST00000515021.1 ENST00000510490.1 ENST00000316423.6 |
UGDH |
UDP-glucose 6-dehydrogenase |
| chr3_-_145878954 | 0.10 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_-_172198190 | 0.09 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr6_+_108882069 | 0.09 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr22_-_36903101 | 0.09 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr14_-_88459182 | 0.09 |
ENST00000544807.2 |
GALC |
galactosylceramidase |
| chr14_-_88459503 | 0.09 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr10_-_44070016 | 0.09 |
ENST00000374446.2 ENST00000426961.1 ENST00000535642.1 |
ZNF239 |
zinc finger protein 239 |
| chr12_+_7023735 | 0.09 |
ENST00000538763.1 ENST00000544774.1 ENST00000545045.2 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr12_+_7023491 | 0.09 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr22_-_36903069 | 0.08 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr12_+_54348618 | 0.07 |
ENST00000243103.3 |
HOXC12 |
homeobox C12 |
| chr5_-_146889619 | 0.07 |
ENST00000343218.5 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr17_-_42907564 | 0.07 |
ENST00000592524.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr3_-_52719810 | 0.07 |
ENST00000424867.1 ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1 |
polybromo 1 |
| chr1_-_159894319 | 0.07 |
ENST00000320307.4 |
TAGLN2 |
transgelin 2 |
| chr11_-_118901559 | 0.07 |
ENST00000330775.7 ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr17_-_42908155 | 0.06 |
ENST00000426548.1 ENST00000590758.1 ENST00000591424.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr16_+_66878814 | 0.06 |
ENST00000394069.3 |
CA7 |
carbonic anhydrase VII |
| chr17_-_67323305 | 0.06 |
ENST00000392677.2 ENST00000593153.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr17_-_67323232 | 0.06 |
ENST00000592568.1 ENST00000392676.3 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr4_+_129732467 | 0.06 |
ENST00000413543.2 |
PHF17 |
jade family PHD finger 1 |
| chr1_-_241520525 | 0.06 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr7_-_127032363 | 0.06 |
ENST00000393312.1 |
ZNF800 |
zinc finger protein 800 |
| chr6_+_151561085 | 0.06 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr5_-_159797627 | 0.06 |
ENST00000393975.3 |
C1QTNF2 |
C1q and tumor necrosis factor related protein 2 |
| chr20_+_33464238 | 0.05 |
ENST00000360596.2 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr11_+_12132117 | 0.05 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr3_-_127541679 | 0.05 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr20_+_33464407 | 0.05 |
ENST00000253382.5 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr13_-_44361025 | 0.05 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chr9_+_4490394 | 0.05 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr4_-_104119528 | 0.05 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chrX_-_57147748 | 0.05 |
ENST00000374910.3 |
SPIN2B |
spindlin family, member 2B |
| chrX_-_13956737 | 0.05 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chr2_+_170683942 | 0.05 |
ENST00000272793.5 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr2_+_170683979 | 0.05 |
ENST00000418381.1 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr9_-_110251836 | 0.05 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr6_-_153304148 | 0.05 |
ENST00000229758.3 |
FBXO5 |
F-box protein 5 |
| chr17_-_74303761 | 0.05 |
ENST00000262765.5 |
QRICH2 |
glutamine rich 2 |
| chr20_+_56964169 | 0.05 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_126112794 | 0.05 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr20_+_30946106 | 0.05 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chrX_+_69509927 | 0.05 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chrX_-_13956497 | 0.05 |
ENST00000398361.3 |
GPM6B |
glycoprotein M6B |
| chr14_-_24685246 | 0.05 |
ENST00000396833.2 ENST00000288087.7 |
MDP1 |
magnesium-dependent phosphatase 1 |
| chr16_-_4897266 | 0.04 |
ENST00000591451.1 ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr14_-_23770683 | 0.04 |
ENST00000561437.1 ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E |
protein phosphatase 1, regulatory subunit 3E |
| chr6_+_151561506 | 0.04 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr17_-_8059638 | 0.04 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chr12_+_58013693 | 0.04 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr17_-_8534031 | 0.04 |
ENST00000411957.1 ENST00000396239.1 ENST00000379980.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr3_-_50374869 | 0.04 |
ENST00000327761.3 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr1_-_26232951 | 0.04 |
ENST00000426559.2 ENST00000455785.2 |
STMN1 |
stathmin 1 |
| chr11_+_74204395 | 0.04 |
ENST00000526036.1 |
AP001372.2 |
AP001372.2 |
| chr17_-_8534067 | 0.04 |
ENST00000360416.3 ENST00000269243.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr22_-_31741757 | 0.04 |
ENST00000215919.3 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr20_-_17662705 | 0.04 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chrX_-_57164058 | 0.04 |
ENST00000374906.3 |
SPIN2A |
spindlin family, member 2A |
| chr16_+_67198683 | 0.04 |
ENST00000517685.1 ENST00000521374.1 ENST00000584272.1 |
HSF4 |
heat shock transcription factor 4 |
| chr1_+_110162448 | 0.04 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr20_-_3065362 | 0.04 |
ENST00000380293.3 |
AVP |
arginine vasopressin |
| chrX_-_34675391 | 0.04 |
ENST00000275954.3 |
TMEM47 |
transmembrane protein 47 |
| chr17_+_42148097 | 0.04 |
ENST00000269097.4 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
| chr2_+_10183651 | 0.04 |
ENST00000305883.1 |
KLF11 |
Kruppel-like factor 11 |
| chr16_+_16043406 | 0.03 |
ENST00000399410.3 ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr1_-_155826967 | 0.03 |
ENST00000368331.1 ENST00000361040.5 ENST00000271883.5 |
GON4L |
gon-4-like (C. elegans) |
| chr16_+_28835437 | 0.03 |
ENST00000568266.1 |
ATXN2L |
ataxin 2-like |
| chr4_-_186317034 | 0.03 |
ENST00000505916.1 |
LRP2BP |
LRP2 binding protein |
| chr20_+_42086525 | 0.03 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr7_-_92465868 | 0.03 |
ENST00000424848.2 |
CDK6 |
cyclin-dependent kinase 6 |
| chr13_-_107220455 | 0.03 |
ENST00000400198.3 |
ARGLU1 |
arginine and glutamate rich 1 |
| chr12_-_48213568 | 0.03 |
ENST00000080059.7 ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7 |
histone deacetylase 7 |
| chr3_-_183543301 | 0.03 |
ENST00000318631.3 ENST00000431348.1 |
MAP6D1 |
MAP6 domain containing 1 |
| chr2_+_204192942 | 0.03 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr20_-_17662878 | 0.03 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr4_+_17578815 | 0.03 |
ENST00000226299.4 |
LAP3 |
leucine aminopeptidase 3 |
| chr11_-_57283159 | 0.03 |
ENST00000533263.1 ENST00000278426.3 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr3_-_71632894 | 0.03 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chr7_-_2595337 | 0.03 |
ENST00000340611.4 |
BRAT1 |
BRCA1-associated ATM activator 1 |
| chr2_-_220083076 | 0.03 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr17_-_27053216 | 0.03 |
ENST00000292090.3 |
TLCD1 |
TLC domain containing 1 |
| chr11_+_6624955 | 0.03 |
ENST00000299421.4 ENST00000537806.1 |
ILK |
integrin-linked kinase |
| chr16_+_447209 | 0.03 |
ENST00000382940.4 ENST00000219479.2 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr11_+_6625046 | 0.03 |
ENST00000396751.2 |
ILK |
integrin-linked kinase |
| chr17_+_76311791 | 0.03 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr1_-_26233423 | 0.03 |
ENST00000357865.2 |
STMN1 |
stathmin 1 |
| chr17_+_25621102 | 0.03 |
ENST00000581440.1 ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1 |
WD repeat and SOCS box containing 1 |
| chr6_-_153304697 | 0.03 |
ENST00000367241.3 |
FBXO5 |
F-box protein 5 |
| chr11_+_6624970 | 0.03 |
ENST00000420936.2 ENST00000528995.1 |
ILK |
integrin-linked kinase |
| chr2_+_28113583 | 0.03 |
ENST00000344773.2 ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE |
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr5_-_131563501 | 0.03 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr7_+_44084233 | 0.03 |
ENST00000448521.1 |
DBNL |
drebrin-like |
| chr16_+_4897912 | 0.03 |
ENST00000545171.1 |
UBN1 |
ubinuclein 1 |
| chrX_-_57147902 | 0.03 |
ENST00000275988.5 ENST00000434397.1 ENST00000333933.3 ENST00000374912.5 |
SPIN2B |
spindlin family, member 2B |
| chr1_-_8939265 | 0.03 |
ENST00000489867.1 |
ENO1 |
enolase 1, (alpha) |
| chr1_-_26232522 | 0.03 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr12_-_121734489 | 0.03 |
ENST00000412367.2 ENST00000402834.4 ENST00000404169.3 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr9_-_139922726 | 0.03 |
ENST00000265662.5 ENST00000371605.3 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr2_-_73460334 | 0.03 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr15_+_38544476 | 0.03 |
ENST00000299084.4 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
| chr9_-_115983641 | 0.03 |
ENST00000238256.3 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr17_+_66508537 | 0.03 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_-_2614927 | 0.03 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr5_-_150138551 | 0.03 |
ENST00000446090.2 ENST00000447998.2 |
DCTN4 |
dynactin 4 (p62) |
| chr15_-_34875771 | 0.03 |
ENST00000267731.7 |
GOLGA8B |
golgin A8 family, member B |
| chr16_+_4897632 | 0.03 |
ENST00000262376.6 |
UBN1 |
ubinuclein 1 |
| chr5_-_171881491 | 0.03 |
ENST00000311601.5 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr16_+_53164833 | 0.03 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr9_-_115983568 | 0.02 |
ENST00000446284.1 ENST00000414250.1 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr6_-_43021612 | 0.02 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
| chr12_+_50355647 | 0.02 |
ENST00000293599.6 |
AQP5 |
aquaporin 5 |
| chr17_-_73127353 | 0.02 |
ENST00000580423.1 ENST00000578337.1 ENST00000582160.1 |
NT5C |
5', 3'-nucleotidase, cytosolic |
| chr1_+_45805342 | 0.02 |
ENST00000372090.5 |
TOE1 |
target of EGR1, member 1 (nuclear) |
| chr22_-_36424458 | 0.02 |
ENST00000438146.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_88178964 | 0.02 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr18_+_43913919 | 0.02 |
ENST00000587853.1 |
RNF165 |
ring finger protein 165 |
| chr5_+_138609441 | 0.02 |
ENST00000509990.1 ENST00000506147.1 ENST00000512107.1 |
MATR3 |
matrin 3 |
| chr17_-_35969409 | 0.02 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr17_+_42148225 | 0.02 |
ENST00000591696.1 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
| chr14_-_20923195 | 0.02 |
ENST00000206542.4 |
OSGEP |
O-sialoglycoprotein endopeptidase |
| chr3_-_81811312 | 0.02 |
ENST00000429644.2 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr2_-_28113217 | 0.02 |
ENST00000444339.2 |
RBKS |
ribokinase |
| chr4_+_186317133 | 0.02 |
ENST00000507753.1 |
ANKRD37 |
ankyrin repeat domain 37 |
| chr19_-_44259136 | 0.02 |
ENST00000270066.6 |
SMG9 |
SMG9 nonsense mediated mRNA decay factor |
| chr19_+_4472230 | 0.02 |
ENST00000301284.4 ENST00000586684.1 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
| chrX_-_57163430 | 0.02 |
ENST00000374908.1 |
SPIN2A |
spindlin family, member 2A |
| chr11_+_76156045 | 0.02 |
ENST00000533988.1 ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30 |
chromosome 11 open reading frame 30 |
| chr17_-_2206801 | 0.02 |
ENST00000544865.1 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr3_+_49507559 | 0.02 |
ENST00000421560.1 ENST00000308775.2 ENST00000545947.1 ENST00000541308.1 ENST00000539901.1 ENST00000538711.1 ENST00000418588.1 |
DAG1 |
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr16_+_447226 | 0.02 |
ENST00000433358.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr1_-_43833628 | 0.02 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr13_+_115079949 | 0.02 |
ENST00000361283.1 |
CHAMP1 |
chromosome alignment maintaining phosphoprotein 1 |
| chr5_-_88179017 | 0.02 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr21_-_38639601 | 0.02 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr5_+_134094461 | 0.02 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr19_-_55691614 | 0.02 |
ENST00000592470.1 ENST00000354308.3 |
SYT5 |
synaptotagmin V |
| chr6_+_87865262 | 0.02 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr3_+_49044798 | 0.02 |
ENST00000438660.1 ENST00000608424.1 ENST00000415265.2 |
WDR6 |
WD repeat domain 6 |
| chr5_+_49962495 | 0.02 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr7_+_44084262 | 0.02 |
ENST00000456905.1 ENST00000440166.1 ENST00000452943.1 ENST00000468694.1 ENST00000494774.1 ENST00000490734.2 |
DBNL |
drebrin-like |
| chr5_+_55033845 | 0.02 |
ENST00000353507.5 ENST00000514278.2 ENST00000505374.1 ENST00000506511.1 |
DDX4 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr3_-_49377499 | 0.02 |
ENST00000265560.4 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr22_-_31742218 | 0.02 |
ENST00000266269.5 ENST00000405309.3 ENST00000351933.4 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr6_-_159240415 | 0.02 |
ENST00000367075.3 |
EZR |
ezrin |
| chr9_-_139922631 | 0.02 |
ENST00000341511.6 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr12_-_49318715 | 0.02 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr10_-_101945771 | 0.02 |
ENST00000370408.2 ENST00000407654.3 |
ERLIN1 |
ER lipid raft associated 1 |
| chr9_+_132388423 | 0.02 |
ENST00000372483.4 ENST00000459968.2 ENST00000482347.1 ENST00000372481.3 |
NTMT1 |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr10_-_22292675 | 0.02 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr8_+_55047763 | 0.02 |
ENST00000260102.4 ENST00000519831.1 |
MRPL15 |
mitochondrial ribosomal protein L15 |
| chr7_+_100318423 | 0.02 |
ENST00000252723.2 |
EPO |
erythropoietin |
| chr16_+_2098003 | 0.02 |
ENST00000439673.2 ENST00000350773.4 |
TSC2 |
tuberous sclerosis 2 |
| chr19_+_18942720 | 0.02 |
ENST00000262803.5 |
UPF1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr4_+_4291924 | 0.02 |
ENST00000355834.3 ENST00000337872.4 ENST00000538529.1 ENST00000502918.1 |
ZBTB49 |
zinc finger and BTB domain containing 49 |
| chr12_+_110906169 | 0.02 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr14_+_64970662 | 0.02 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr13_-_28024681 | 0.02 |
ENST00000381116.1 ENST00000381120.3 ENST00000431572.2 |
MTIF3 |
mitochondrial translational initiation factor 3 |
| chr17_-_73127826 | 0.02 |
ENST00000582170.1 ENST00000245552.2 |
NT5C |
5', 3'-nucleotidase, cytosolic |
| chr13_-_31038370 | 0.02 |
ENST00000399489.1 ENST00000339872.4 |
HMGB1 |
high mobility group box 1 |
| chr11_-_64014379 | 0.02 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr8_+_95908041 | 0.02 |
ENST00000396113.1 ENST00000519136.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr11_+_33279850 | 0.02 |
ENST00000531504.1 ENST00000456517.1 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr18_+_77439775 | 0.02 |
ENST00000299543.7 ENST00000075430.7 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr1_-_51425772 | 0.02 |
ENST00000371778.4 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr1_+_204797749 | 0.02 |
ENST00000367172.4 ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC |
neurofascin |
| chr1_-_166944561 | 0.02 |
ENST00000271417.3 |
ILDR2 |
immunoglobulin-like domain containing receptor 2 |
| chr22_-_21356375 | 0.02 |
ENST00000215742.4 ENST00000399133.2 |
THAP7 |
THAP domain containing 7 |
| chr2_+_242254507 | 0.02 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chrX_-_102941596 | 0.02 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr10_-_16859361 | 0.02 |
ENST00000377921.3 |
RSU1 |
Ras suppressor protein 1 |
| chr10_-_75910789 | 0.02 |
ENST00000355264.4 |
AP3M1 |
adaptor-related protein complex 3, mu 1 subunit |
| chr10_-_16859442 | 0.02 |
ENST00000602389.1 ENST00000345264.5 |
RSU1 |
Ras suppressor protein 1 |
| chr9_+_133884469 | 0.02 |
ENST00000361069.4 |
LAMC3 |
laminin, gamma 3 |
| chr2_+_178077477 | 0.02 |
ENST00000411529.2 ENST00000435711.1 |
HNRNPA3 |
heterogeneous nuclear ribonucleoprotein A3 |
| chr10_+_93558069 | 0.02 |
ENST00000371627.4 |
TNKS2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr19_+_1407733 | 0.01 |
ENST00000592453.1 |
DAZAP1 |
DAZ associated protein 1 |
| chr2_-_216946500 | 0.01 |
ENST00000265322.7 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
| chr20_+_56964253 | 0.01 |
ENST00000395802.3 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_49962772 | 0.01 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr10_+_122216316 | 0.01 |
ENST00000398250.1 ENST00000439221.1 ENST00000398248.1 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr1_+_193091080 | 0.01 |
ENST00000367435.3 |
CDC73 |
cell division cycle 73 |
| chr10_+_180987 | 0.01 |
ENST00000381591.1 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr15_-_43622736 | 0.01 |
ENST00000544735.1 ENST00000567039.1 ENST00000305641.5 |
LCMT2 |
leucine carboxyl methyltransferase 2 |
| chr12_+_6643676 | 0.01 |
ENST00000396856.1 ENST00000396861.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_8938736 | 0.01 |
ENST00000234590.4 |
ENO1 |
enolase 1, (alpha) |
| chr9_+_115983808 | 0.01 |
ENST00000374210.6 ENST00000374212.4 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
| chr12_-_2113583 | 0.01 |
ENST00000397173.4 ENST00000280665.6 |
DCP1B |
decapping mRNA 1B |
| chr19_-_1592828 | 0.01 |
ENST00000592012.1 |
MBD3 |
methyl-CpG binding domain protein 3 |
| chr4_-_25161996 | 0.01 |
ENST00000513285.1 ENST00000382103.2 |
SEPSECS |
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr19_-_1592652 | 0.01 |
ENST00000156825.1 ENST00000434436.3 |
MBD3 |
methyl-CpG binding domain protein 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |