Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
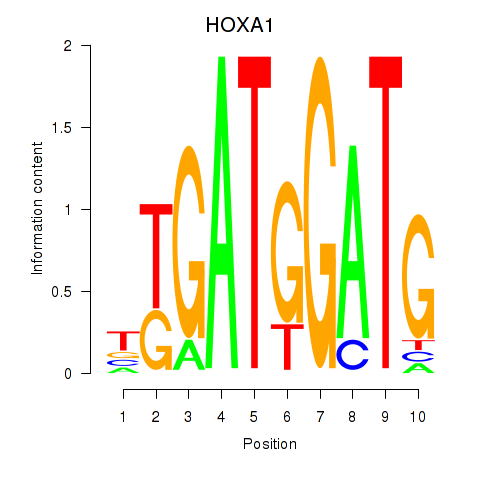
Results for HOXA1
Z-value: 0.29
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.7 | HOXA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg19_v2_chr7_-_27135591_27135658 | 0.38 | 3.5e-01 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10697895 | 1.39 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr1_+_183155373 | 0.36 |
ENST00000493293.1 ENST00000264144.4 |
LAMC2 |
laminin, gamma 2 |
| chr5_+_167181917 | 0.36 |
ENST00000519204.1 |
TENM2 |
teneurin transmembrane protein 2 |
| chr19_+_54371114 | 0.36 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr12_-_85306562 | 0.32 |
ENST00000551612.1 ENST00000450363.3 ENST00000552192.1 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr16_+_71660079 | 0.31 |
ENST00000565261.1 ENST00000268485.3 ENST00000299952.4 |
MARVELD3 |
MARVEL domain containing 3 |
| chr16_+_71660052 | 0.30 |
ENST00000567566.1 |
MARVELD3 |
MARVEL domain containing 3 |
| chr12_-_95510743 | 0.30 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr4_+_113970772 | 0.29 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr1_+_2005425 | 0.28 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr8_+_37887772 | 0.24 |
ENST00000338825.4 |
EIF4EBP1 |
eukaryotic translation initiation factor 4E binding protein 1 |
| chr12_-_85306594 | 0.22 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr3_-_112564797 | 0.22 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr1_+_86889769 | 0.21 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr7_-_22233442 | 0.21 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr20_-_30458491 | 0.21 |
ENST00000339738.5 |
DUSP15 |
dual specificity phosphatase 15 |
| chr16_+_75256507 | 0.20 |
ENST00000495583.1 |
CTRB1 |
chymotrypsinogen B1 |
| chr2_+_65215604 | 0.20 |
ENST00000531327.1 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr10_-_105615164 | 0.19 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr6_+_80129989 | 0.19 |
ENST00000429444.1 |
RP1-232L24.3 |
RP1-232L24.3 |
| chr1_+_107682629 | 0.18 |
ENST00000370074.4 ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1 |
netrin G1 |
| chr1_+_107683436 | 0.17 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr1_+_205473720 | 0.17 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr11_-_72353451 | 0.17 |
ENST00000376450.3 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr6_+_90272027 | 0.16 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr2_-_75788038 | 0.16 |
ENST00000393913.3 ENST00000410113.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr9_-_5304432 | 0.16 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr2_+_1488435 | 0.16 |
ENST00000446278.1 ENST00000469607.1 |
TPO |
thyroid peroxidase |
| chr16_+_23847267 | 0.15 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr20_+_52105495 | 0.15 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr2_-_71062938 | 0.14 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr12_-_52911718 | 0.14 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr15_+_48009541 | 0.14 |
ENST00000536845.2 ENST00000558816.1 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr19_-_38746979 | 0.12 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr6_+_121756809 | 0.12 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr15_-_63674034 | 0.12 |
ENST00000344366.3 ENST00000422263.2 |
CA12 |
carbonic anhydrase XII |
| chr5_+_54398463 | 0.12 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr6_+_31021225 | 0.12 |
ENST00000565192.1 ENST00000562344.1 |
HCG22 |
HLA complex group 22 |
| chr5_+_36152179 | 0.12 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr15_+_84908573 | 0.12 |
ENST00000424966.1 ENST00000422563.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr10_+_98741041 | 0.11 |
ENST00000286067.2 |
C10orf12 |
chromosome 10 open reading frame 12 |
| chr12_+_75728419 | 0.11 |
ENST00000378695.4 ENST00000312442.2 |
GLIPR1L1 |
GLI pathogenesis-related 1 like 1 |
| chr15_-_63674218 | 0.10 |
ENST00000178638.3 |
CA12 |
carbonic anhydrase XII |
| chr5_+_36152091 | 0.10 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr7_-_994302 | 0.10 |
ENST00000265846.5 |
ADAP1 |
ArfGAP with dual PH domains 1 |
| chr8_-_91095099 | 0.10 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr17_-_8027402 | 0.10 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr9_+_124329336 | 0.10 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr8_+_1993173 | 0.10 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr6_+_31021973 | 0.10 |
ENST00000570223.1 ENST00000566475.1 ENST00000426185.1 |
HCG22 |
HLA complex group 22 |
| chr1_-_24469602 | 0.10 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr7_-_101212244 | 0.10 |
ENST00000451953.1 ENST00000434537.1 ENST00000437900.1 |
LINC01007 |
long intergenic non-protein coding RNA 1007 |
| chr7_-_100808394 | 0.10 |
ENST00000445482.2 |
VGF |
VGF nerve growth factor inducible |
| chr7_-_100808843 | 0.09 |
ENST00000249330.2 |
VGF |
VGF nerve growth factor inducible |
| chr19_-_51141196 | 0.09 |
ENST00000338916.4 |
SYT3 |
synaptotagmin III |
| chrX_+_82763265 | 0.09 |
ENST00000373200.2 |
POU3F4 |
POU class 3 homeobox 4 |
| chrX_+_130192318 | 0.09 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr18_+_11752040 | 0.09 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_-_153513170 | 0.09 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr8_+_1993152 | 0.09 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr21_-_46340884 | 0.08 |
ENST00000302347.5 ENST00000517819.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_+_36152163 | 0.08 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_+_86099884 | 0.08 |
ENST00000517476.1 ENST00000521429.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr9_+_113431059 | 0.08 |
ENST00000416899.2 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr3_+_14444063 | 0.08 |
ENST00000454876.2 ENST00000360861.3 ENST00000416216.2 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr17_-_15501932 | 0.08 |
ENST00000583965.1 |
CDRT1 |
CMT1A duplicated region transcript 1 |
| chr19_-_42927251 | 0.08 |
ENST00000597001.1 |
LIPE |
lipase, hormone-sensitive |
| chr18_-_74207146 | 0.08 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr7_+_79765071 | 0.08 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr1_+_45140360 | 0.08 |
ENST00000418644.1 ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228 |
chromosome 1 open reading frame 228 |
| chr15_+_71839566 | 0.07 |
ENST00000357769.4 |
THSD4 |
thrombospondin, type I, domain containing 4 |
| chr7_-_3214287 | 0.07 |
ENST00000404626.3 |
AC091801.1 |
LOC392621; Uncharacterized protein |
| chr1_+_12916941 | 0.07 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr4_-_123542224 | 0.07 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr8_-_119124045 | 0.07 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr9_+_125281420 | 0.07 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr4_-_90229142 | 0.07 |
ENST00000609438.1 |
GPRIN3 |
GPRIN family member 3 |
| chr9_+_113431029 | 0.07 |
ENST00000189978.5 ENST00000374448.4 ENST00000374440.3 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr16_+_30034655 | 0.07 |
ENST00000300575.2 |
C16orf92 |
chromosome 16 open reading frame 92 |
| chr20_+_31669333 | 0.07 |
ENST00000375483.3 |
BPIFB4 |
BPI fold containing family B, member 4 |
| chr14_-_77965151 | 0.07 |
ENST00000393684.3 ENST00000493585.1 ENST00000554801.2 ENST00000342219.4 ENST00000412904.1 ENST00000429906.1 |
ISM2 |
isthmin 2 |
| chr3_+_132757215 | 0.07 |
ENST00000321871.6 ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108 |
transmembrane protein 108 |
| chr11_+_61248583 | 0.07 |
ENST00000432063.2 ENST00000338608.2 |
PPP1R32 |
protein phosphatase 1, regulatory subunit 32 |
| chr7_+_139529040 | 0.06 |
ENST00000455353.1 ENST00000458722.1 ENST00000411653.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr19_-_51192661 | 0.06 |
ENST00000391813.1 |
SHANK1 |
SH3 and multiple ankyrin repeat domains 1 |
| chr7_+_97736197 | 0.06 |
ENST00000297293.5 |
LMTK2 |
lemur tyrosine kinase 2 |
| chr7_+_139528952 | 0.06 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr19_+_5681153 | 0.06 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr6_-_154677900 | 0.06 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr3_-_113918254 | 0.06 |
ENST00000460779.1 |
DRD3 |
dopamine receptor D3 |
| chr14_+_24779340 | 0.06 |
ENST00000533293.1 ENST00000543919.1 |
LTB4R2 |
leukotriene B4 receptor 2 |
| chr19_+_19144384 | 0.06 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr9_-_130679257 | 0.06 |
ENST00000361444.3 ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr11_+_7626950 | 0.06 |
ENST00000530181.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_11450249 | 0.06 |
ENST00000222120.3 |
RAB3D |
RAB3D, member RAS oncogene family |
| chr2_-_49381572 | 0.06 |
ENST00000454032.1 ENST00000304421.4 |
FSHR |
follicle stimulating hormone receptor |
| chr2_-_49381646 | 0.06 |
ENST00000346173.3 ENST00000406846.2 |
FSHR |
follicle stimulating hormone receptor |
| chr1_+_165864800 | 0.06 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chrX_+_139791917 | 0.06 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chrX_+_37639302 | 0.06 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr17_-_27278445 | 0.05 |
ENST00000268756.3 ENST00000584685.1 |
PHF12 |
PHD finger protein 12 |
| chr7_+_20370746 | 0.05 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr21_-_32185570 | 0.05 |
ENST00000329621.4 |
KRTAP8-1 |
keratin associated protein 8-1 |
| chr6_+_73331520 | 0.05 |
ENST00000342056.2 ENST00000355194.4 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr6_+_24775641 | 0.05 |
ENST00000378054.2 ENST00000476555.1 |
GMNN |
geminin, DNA replication inhibitor |
| chr5_-_137071689 | 0.05 |
ENST00000505853.1 |
KLHL3 |
kelch-like family member 3 |
| chrX_+_128913906 | 0.05 |
ENST00000356892.3 |
SASH3 |
SAM and SH3 domain containing 3 |
| chr7_-_44180884 | 0.05 |
ENST00000458240.1 ENST00000223364.3 |
MYL7 |
myosin, light chain 7, regulatory |
| chr1_-_207095324 | 0.05 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr16_-_55867146 | 0.05 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr4_+_177241094 | 0.05 |
ENST00000503362.1 |
SPCS3 |
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr7_+_23637763 | 0.05 |
ENST00000410069.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr16_+_23847339 | 0.05 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr13_-_20077417 | 0.05 |
ENST00000382978.1 ENST00000400230.2 ENST00000255310.6 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr1_-_201096312 | 0.05 |
ENST00000449188.2 |
ASCL5 |
achaete-scute family bHLH transcription factor 5 |
| chr1_-_205391178 | 0.05 |
ENST00000367153.4 ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1 |
LEM domain containing 1 |
| chr16_-_9030515 | 0.05 |
ENST00000535863.1 ENST00000381886.4 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chrX_+_150148976 | 0.05 |
ENST00000419110.1 |
HMGB3 |
high mobility group box 3 |
| chr20_-_30458432 | 0.05 |
ENST00000375966.4 ENST00000278979.3 |
DUSP15 |
dual specificity phosphatase 15 |
| chr10_+_7745303 | 0.05 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr16_+_29674540 | 0.04 |
ENST00000436527.1 ENST00000360121.3 ENST00000449759.1 |
SPN QPRT |
sialophorin quinolinate phosphoribosyltransferase |
| chr14_-_81408093 | 0.04 |
ENST00000555265.1 |
CEP128 |
centrosomal protein 128kDa |
| chr10_+_62538089 | 0.04 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr8_+_98788003 | 0.04 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr17_+_60447579 | 0.04 |
ENST00000450662.2 |
EFCAB3 |
EF-hand calcium binding domain 3 |
| chr11_+_61717279 | 0.04 |
ENST00000378043.4 |
BEST1 |
bestrophin 1 |
| chr1_-_113498943 | 0.04 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr5_+_63802109 | 0.04 |
ENST00000334025.2 |
RGS7BP |
regulator of G-protein signaling 7 binding protein |
| chr20_+_44036620 | 0.04 |
ENST00000372710.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_+_100136811 | 0.04 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr11_+_61717336 | 0.04 |
ENST00000378042.3 |
BEST1 |
bestrophin 1 |
| chr9_-_123676827 | 0.04 |
ENST00000546084.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr11_-_2162162 | 0.04 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr19_-_33360647 | 0.04 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr1_-_207095212 | 0.04 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr11_+_46193466 | 0.04 |
ENST00000533793.1 |
RP11-702F3.3 |
RP11-702F3.3 |
| chr12_-_101604185 | 0.04 |
ENST00000536262.2 |
SLC5A8 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr17_-_38821373 | 0.04 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr13_-_20735178 | 0.04 |
ENST00000241125.3 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
| chr20_+_44036900 | 0.04 |
ENST00000443296.1 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_+_46152886 | 0.04 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr6_-_142409936 | 0.04 |
ENST00000258042.1 |
NMBR |
neuromedin B receptor |
| chr12_-_86650045 | 0.04 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_+_8276495 | 0.04 |
ENST00000546339.1 |
CLEC4A |
C-type lectin domain family 4, member A |
| chr11_+_68228186 | 0.04 |
ENST00000393799.2 ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
| chr12_+_57998400 | 0.04 |
ENST00000548804.1 ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr3_-_161090660 | 0.04 |
ENST00000359175.4 |
SPTSSB |
serine palmitoyltransferase, small subunit B |
| chr10_+_7745232 | 0.04 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_+_66247478 | 0.04 |
ENST00000531863.1 ENST00000532677.1 |
DPP3 |
dipeptidyl-peptidase 3 |
| chr9_-_97402531 | 0.04 |
ENST00000415431.1 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
| chr10_+_62538248 | 0.04 |
ENST00000448257.2 |
CDK1 |
cyclin-dependent kinase 1 |
| chr12_+_125478241 | 0.04 |
ENST00000341446.8 |
BRI3BP |
BRI3 binding protein |
| chr6_-_44233361 | 0.04 |
ENST00000275015.5 |
NFKBIE |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr14_-_81408031 | 0.04 |
ENST00000216517.6 |
CEP128 |
centrosomal protein 128kDa |
| chr17_-_73401567 | 0.03 |
ENST00000392562.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr18_-_3874271 | 0.03 |
ENST00000400149.3 ENST00000400155.1 ENST00000400150.3 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr10_-_126849068 | 0.03 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr3_+_48264816 | 0.03 |
ENST00000296435.2 ENST00000576243.1 |
CAMP |
cathelicidin antimicrobial peptide |
| chr11_+_2482661 | 0.03 |
ENST00000335475.5 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr6_-_149867122 | 0.03 |
ENST00000253329.2 |
PPIL4 |
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr19_+_54641444 | 0.03 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr16_-_52119019 | 0.03 |
ENST00000561513.1 ENST00000565742.1 |
LINC00919 |
long intergenic non-protein coding RNA 919 |
| chr9_-_97402413 | 0.03 |
ENST00000414122.1 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
| chr9_+_116037922 | 0.03 |
ENST00000374198.4 |
PRPF4 |
pre-mRNA processing factor 4 |
| chr22_+_39101728 | 0.03 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr1_+_240255166 | 0.03 |
ENST00000319653.9 |
FMN2 |
formin 2 |
| chr1_-_217804377 | 0.03 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr17_+_43239191 | 0.03 |
ENST00000589230.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr8_-_101734907 | 0.03 |
ENST00000318607.5 ENST00000521865.1 ENST00000520804.1 ENST00000522720.1 ENST00000521067.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr3_+_111260856 | 0.03 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr4_-_38806404 | 0.03 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr12_+_10658489 | 0.03 |
ENST00000538173.1 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr17_+_79495397 | 0.03 |
ENST00000417245.2 ENST00000334850.7 |
FSCN2 |
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr17_-_3499125 | 0.03 |
ENST00000399759.3 |
TRPV1 |
transient receptor potential cation channel, subfamily V, member 1 |
| chr12_-_121342170 | 0.03 |
ENST00000353487.2 |
SPPL3 |
signal peptide peptidase like 3 |
| chr3_-_129612394 | 0.03 |
ENST00000505616.1 ENST00000426664.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr14_-_81408063 | 0.03 |
ENST00000557411.1 |
CEP128 |
centrosomal protein 128kDa |
| chr16_+_66914264 | 0.03 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr11_-_5248294 | 0.03 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr12_+_58148842 | 0.03 |
ENST00000266643.5 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
| chr22_-_22307199 | 0.03 |
ENST00000397495.4 ENST00000263212.5 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr6_+_50061315 | 0.03 |
ENST00000415106.1 |
RP11-397G17.1 |
RP11-397G17.1 |
| chr17_-_44896047 | 0.03 |
ENST00000225512.5 |
WNT3 |
wingless-type MMTV integration site family, member 3 |
| chr10_-_103599591 | 0.03 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr10_-_101380121 | 0.03 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr3_+_111260954 | 0.03 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr15_-_62937380 | 0.03 |
ENST00000560347.1 ENST00000558940.1 |
RP11-625H11.1 |
Uncharacterized protein |
| chr3_+_130150307 | 0.03 |
ENST00000512836.1 |
COL6A5 |
collagen, type VI, alpha 5 |
| chr3_-_150264272 | 0.03 |
ENST00000491660.1 ENST00000487153.1 ENST00000239944.2 |
SERP1 |
stress-associated endoplasmic reticulum protein 1 |
| chr6_-_47009996 | 0.03 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr3_+_44916098 | 0.03 |
ENST00000296125.4 |
TGM4 |
transglutaminase 4 |
| chr18_-_3874247 | 0.03 |
ENST00000581699.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_-_40098672 | 0.02 |
ENST00000535435.1 |
HEYL |
hes-related family bHLH transcription factor with YRPW motif-like |
| chr19_+_1026298 | 0.02 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr3_+_4535025 | 0.02 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr12_-_10962767 | 0.02 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr11_+_60739249 | 0.02 |
ENST00000542157.1 ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6 |
CD6 molecule |
| chr4_+_71200681 | 0.02 |
ENST00000273936.5 |
CABS1 |
calcium-binding protein, spermatid-specific 1 |
| chr2_+_233897382 | 0.02 |
ENST00000233840.3 |
NEU2 |
sialidase 2 (cytosolic sialidase) |
| chr11_-_88796803 | 0.02 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.6 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 1.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.0 | GO:0015811 | L-cystine transport(GO:0015811) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 1.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |