Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
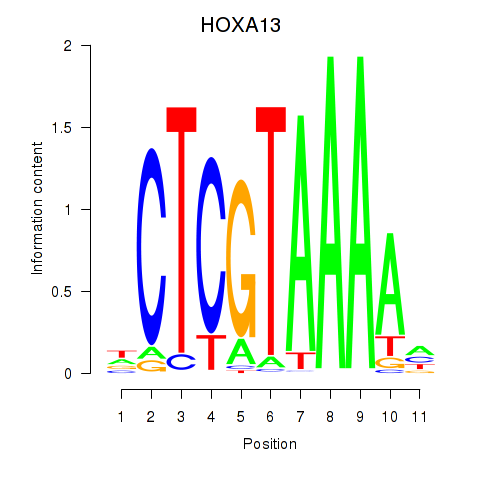
Results for HOXA13
Z-value: 0.58
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | HOXA13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg19_v2_chr7_-_27239703_27239725 | -0.57 | 1.4e-01 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 1.41 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr14_-_92414055 | 1.19 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr2_-_225811747 | 1.00 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr2_+_33359646 | 0.91 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.90 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr12_-_91539918 | 0.85 |
ENST00000548218.1 |
DCN |
decorin |
| chr3_-_100712352 | 0.75 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr5_-_19988339 | 0.63 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr12_-_91546926 | 0.60 |
ENST00000550758.1 |
DCN |
decorin |
| chr1_+_78769549 | 0.60 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr12_+_75874580 | 0.55 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr1_+_78383813 | 0.39 |
ENST00000342754.5 |
NEXN |
nexilin (F actin binding protein) |
| chr17_-_39538550 | 0.38 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr12_+_75874460 | 0.38 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr5_-_146781153 | 0.34 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr18_-_70532906 | 0.32 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr8_-_93029520 | 0.31 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr22_+_38142235 | 0.30 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr21_+_39628655 | 0.29 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_-_149093499 | 0.29 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr15_+_69373184 | 0.27 |
ENST00000558147.1 ENST00000440444.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr7_-_92777606 | 0.26 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr11_-_124670550 | 0.25 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr6_+_73076432 | 0.24 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_-_156877997 | 0.24 |
ENST00000295926.3 |
CCNL1 |
cyclin L1 |
| chr5_-_124081008 | 0.24 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr4_-_128887069 | 0.23 |
ENST00000541133.1 ENST00000296468.3 ENST00000513559.1 |
MFSD8 |
major facilitator superfamily domain containing 8 |
| chr3_-_8811288 | 0.23 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chrX_+_139791917 | 0.22 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr8_-_93029865 | 0.22 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_-_29234802 | 0.22 |
ENST00000449801.1 ENST00000409850.1 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr3_-_123680246 | 0.22 |
ENST00000488653.2 |
CCDC14 |
coiled-coil domain containing 14 |
| chr6_-_51952367 | 0.22 |
ENST00000340994.4 |
PKHD1 |
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr1_-_31845914 | 0.22 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr5_-_138210977 | 0.21 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr2_+_204732666 | 0.21 |
ENST00000295854.6 ENST00000472206.1 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr18_-_10748498 | 0.21 |
ENST00000579949.1 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr8_+_77318769 | 0.20 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr4_-_170533723 | 0.20 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr8_+_133879193 | 0.20 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr12_-_47473425 | 0.20 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr11_+_61891445 | 0.20 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr16_-_90096309 | 0.20 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr3_-_156878540 | 0.20 |
ENST00000461804.1 |
CCNL1 |
cyclin L1 |
| chr1_-_85097431 | 0.19 |
ENST00000327308.3 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr19_-_39694894 | 0.19 |
ENST00000318438.6 |
SYCN |
syncollin |
| chr4_+_70894130 | 0.19 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr10_+_18240834 | 0.19 |
ENST00000377371.3 ENST00000539911.1 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr11_-_104480019 | 0.19 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr19_-_6375860 | 0.19 |
ENST00000245810.1 |
PSPN |
persephin |
| chr3_+_101546827 | 0.19 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr3_-_160823158 | 0.18 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_+_43766642 | 0.18 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr8_+_143781513 | 0.18 |
ENST00000292430.6 ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K |
lymphocyte antigen 6 complex, locus K |
| chr3_-_160823040 | 0.18 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr3_-_150996239 | 0.18 |
ENST00000309170.3 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
| chr15_-_55881135 | 0.18 |
ENST00000302000.6 |
PYGO1 |
pygopus family PHD finger 1 |
| chr17_-_39471947 | 0.18 |
ENST00000334202.3 |
KRTAP17-1 |
keratin associated protein 17-1 |
| chr19_-_44809121 | 0.17 |
ENST00000591609.1 ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235 |
zinc finger protein 235 |
| chr1_+_179050512 | 0.17 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr5_+_169010638 | 0.17 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr10_-_106098162 | 0.17 |
ENST00000337478.1 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr5_-_150284532 | 0.17 |
ENST00000394226.2 ENST00000446148.2 ENST00000274599.5 ENST00000418587.2 |
ZNF300 |
zinc finger protein 300 |
| chr17_-_39637392 | 0.16 |
ENST00000246639.2 ENST00000393989.1 |
KRT35 |
keratin 35 |
| chr2_+_109223595 | 0.16 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr13_+_34392200 | 0.16 |
ENST00000434425.1 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr19_+_13858593 | 0.15 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr22_+_20193905 | 0.15 |
ENST00000609602.1 |
LINC00896 |
long intergenic non-protein coding RNA 896 |
| chr10_+_92631709 | 0.15 |
ENST00000413330.1 ENST00000277882.3 |
RPP30 |
ribonuclease P/MRP 30kDa subunit |
| chr1_+_199996733 | 0.15 |
ENST00000236914.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr17_-_67264947 | 0.15 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr10_+_18240753 | 0.15 |
ENST00000377369.2 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chrX_+_49687216 | 0.15 |
ENST00000376088.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr10_+_18240814 | 0.15 |
ENST00000377374.4 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr3_-_160822858 | 0.15 |
ENST00000488170.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr19_+_16999654 | 0.15 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr10_-_28270795 | 0.15 |
ENST00000545014.1 |
ARMC4 |
armadillo repeat containing 4 |
| chr5_-_150284351 | 0.14 |
ENST00000427179.1 |
ZNF300 |
zinc finger protein 300 |
| chr2_-_86850949 | 0.14 |
ENST00000237455.4 |
RNF103 |
ring finger protein 103 |
| chr14_-_50319482 | 0.14 |
ENST00000546046.1 ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF |
nuclear export mediator factor |
| chrX_+_37850026 | 0.14 |
ENST00000341016.3 |
CXorf27 |
chromosome X open reading frame 27 |
| chr17_+_78234625 | 0.14 |
ENST00000508628.2 ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213 |
ring finger protein 213 |
| chr6_+_131894284 | 0.14 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr3_-_178969403 | 0.14 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr10_+_90750493 | 0.14 |
ENST00000357339.2 ENST00000355279.2 |
FAS |
Fas cell surface death receptor |
| chr5_+_133861790 | 0.13 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr2_-_32390801 | 0.13 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr5_-_138533973 | 0.13 |
ENST00000507002.1 ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1 |
SIL1 nucleotide exchange factor |
| chrX_+_9431324 | 0.13 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr10_-_61513201 | 0.13 |
ENST00000414264.1 ENST00000594536.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr10_-_61513146 | 0.13 |
ENST00000430431.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr10_-_51371321 | 0.13 |
ENST00000602930.1 |
AGAP8 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr1_-_220220000 | 0.13 |
ENST00000366923.3 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr5_+_67584174 | 0.13 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_121131970 | 0.13 |
ENST00000535656.1 |
MLEC |
malectin |
| chr9_-_98269699 | 0.13 |
ENST00000429896.2 |
PTCH1 |
patched 1 |
| chr7_+_95401851 | 0.13 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr11_-_116663127 | 0.12 |
ENST00000433069.1 ENST00000542499.1 |
APOA5 |
apolipoprotein A-V |
| chr8_-_63998590 | 0.12 |
ENST00000260116.4 |
TTPA |
tocopherol (alpha) transfer protein |
| chr16_+_22501658 | 0.12 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr12_-_92539614 | 0.12 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr14_-_94789663 | 0.12 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr20_-_25320367 | 0.12 |
ENST00000450393.1 ENST00000491682.1 |
ABHD12 |
abhydrolase domain containing 12 |
| chr22_+_40342819 | 0.12 |
ENST00000407075.3 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr12_+_120105558 | 0.12 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr19_-_48547294 | 0.12 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chr21_+_39628852 | 0.11 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_24540046 | 0.11 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr1_+_84609944 | 0.11 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_11457162 | 0.11 |
ENST00000590482.1 |
TMEM205 |
transmembrane protein 205 |
| chr1_-_204183071 | 0.11 |
ENST00000308302.3 |
GOLT1A |
golgi transport 1A |
| chr11_+_122709200 | 0.11 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr14_-_50319758 | 0.11 |
ENST00000298310.5 |
NEMF |
nuclear export mediator factor |
| chr2_+_32390925 | 0.11 |
ENST00000440718.1 ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6 |
solute carrier family 30 (zinc transporter), member 6 |
| chr10_-_52383644 | 0.11 |
ENST00000361781.2 |
SGMS1 |
sphingomyelin synthase 1 |
| chr1_-_112106578 | 0.11 |
ENST00000369717.4 |
ADORA3 |
adenosine A3 receptor |
| chr2_+_37571717 | 0.11 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr17_-_39526052 | 0.11 |
ENST00000251646.3 |
KRT33B |
keratin 33B |
| chr14_+_100485712 | 0.11 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chrX_-_19688475 | 0.11 |
ENST00000541422.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr10_+_103986085 | 0.11 |
ENST00000370005.3 |
ELOVL3 |
ELOVL fatty acid elongase 3 |
| chrX_-_38186775 | 0.10 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr19_-_11457082 | 0.10 |
ENST00000587948.1 |
TMEM205 |
transmembrane protein 205 |
| chr10_-_113943447 | 0.10 |
ENST00000369425.1 ENST00000348367.4 ENST00000423155.1 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_+_50575292 | 0.10 |
ENST00000371821.1 ENST00000371819.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr11_+_111896090 | 0.10 |
ENST00000393051.1 |
DLAT |
dihydrolipoamide S-acetyltransferase |
| chrX_+_100878079 | 0.10 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr2_+_37571845 | 0.10 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr7_-_27196267 | 0.10 |
ENST00000242159.3 |
HOXA7 |
homeobox A7 |
| chr10_-_47239738 | 0.10 |
ENST00000413193.2 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr7_-_3214287 | 0.10 |
ENST00000404626.3 |
AC091801.1 |
LOC392621; Uncharacterized protein |
| chr3_+_46618727 | 0.10 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr12_-_111358372 | 0.09 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr6_+_44310376 | 0.09 |
ENST00000515220.1 ENST00000323108.8 |
SPATS1 |
spermatogenesis associated, serine-rich 1 |
| chr12_-_4647606 | 0.09 |
ENST00000261250.3 ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4 |
chromosome 12 open reading frame 4 |
| chr11_-_85565906 | 0.09 |
ENST00000544076.1 |
AP000974.1 |
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
| chr16_-_18812746 | 0.09 |
ENST00000546206.2 ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1 RP11-1035H13.3 |
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr7_+_80275621 | 0.09 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_-_12491608 | 0.09 |
ENST00000545735.1 |
MANSC1 |
MANSC domain containing 1 |
| chr11_+_60995849 | 0.09 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr12_+_119419294 | 0.09 |
ENST00000267260.4 |
SRRM4 |
serine/arginine repetitive matrix 4 |
| chrX_-_135962876 | 0.09 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr6_-_167571817 | 0.08 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr10_+_48189612 | 0.08 |
ENST00000453919.1 |
AGAP9 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr1_+_100818009 | 0.08 |
ENST00000370125.2 ENST00000361544.6 ENST00000370124.3 |
CDC14A |
cell division cycle 14A |
| chr7_+_80275752 | 0.08 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr19_-_51893827 | 0.08 |
ENST00000574814.1 |
CTD-2616J11.4 |
chromosome 19 open reading frame 84 |
| chr15_+_71228826 | 0.08 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr3_+_129159039 | 0.08 |
ENST00000507564.1 ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr17_-_39553844 | 0.08 |
ENST00000251645.2 |
KRT31 |
keratin 31 |
| chr7_-_144107320 | 0.08 |
ENST00000483238.1 ENST00000467773.1 |
NOBOX |
NOBOX oogenesis homeobox |
| chr12_-_71182695 | 0.08 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr14_-_102706934 | 0.08 |
ENST00000523231.1 ENST00000524370.1 ENST00000517966.1 |
MOK |
MOK protein kinase |
| chr21_-_32253874 | 0.08 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chr14_+_56078695 | 0.08 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr21_+_46066331 | 0.08 |
ENST00000334670.8 |
KRTAP10-11 |
keratin associated protein 10-11 |
| chr16_+_66914264 | 0.08 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr20_+_37554955 | 0.07 |
ENST00000217429.4 |
FAM83D |
family with sequence similarity 83, member D |
| chr1_+_199996702 | 0.07 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr10_-_13544945 | 0.07 |
ENST00000378605.3 ENST00000341083.3 |
BEND7 |
BEN domain containing 7 |
| chr9_+_91933726 | 0.07 |
ENST00000534113.2 |
SECISBP2 |
SECIS binding protein 2 |
| chr1_-_28177255 | 0.07 |
ENST00000601459.1 |
AL109927.1 |
HCG2032222; PRO2047; Uncharacterized protein |
| chr4_-_155533787 | 0.07 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr11_+_113779704 | 0.07 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr2_-_178937478 | 0.07 |
ENST00000286063.6 |
PDE11A |
phosphodiesterase 11A |
| chr6_-_114194483 | 0.07 |
ENST00000434296.2 |
RP1-249H1.4 |
RP1-249H1.4 |
| chr17_+_55183261 | 0.07 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr12_-_8693539 | 0.07 |
ENST00000299663.3 |
CLEC4E |
C-type lectin domain family 4, member E |
| chr7_-_130598059 | 0.07 |
ENST00000432045.2 |
MIR29B1 |
microRNA 29a |
| chr7_+_23749894 | 0.07 |
ENST00000433467.2 |
STK31 |
serine/threonine kinase 31 |
| chr16_-_18908196 | 0.07 |
ENST00000565324.1 ENST00000561947.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_8321882 | 0.06 |
ENST00000509453.1 ENST00000503186.1 |
RP11-774O3.2 RP11-774O3.1 |
RP11-774O3.2 RP11-774O3.1 |
| chr20_+_44420617 | 0.06 |
ENST00000449078.1 ENST00000456939.1 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr11_+_101785727 | 0.06 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr6_+_71377473 | 0.06 |
ENST00000370452.3 ENST00000316999.5 ENST00000370455.3 |
SMAP1 |
small ArfGAP 1 |
| chrX_+_117480036 | 0.06 |
ENST00000371822.5 ENST00000254029.3 ENST00000371825.3 |
WDR44 |
WD repeat domain 44 |
| chr19_+_16830774 | 0.06 |
ENST00000524140.2 |
NWD1 |
NACHT and WD repeat domain containing 1 |
| chr2_-_177502254 | 0.06 |
ENST00000339037.3 |
AC017048.3 |
long intergenic non-protein coding RNA 1116 |
| chr3_-_125775629 | 0.06 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr3_-_123339418 | 0.06 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr4_+_122722466 | 0.06 |
ENST00000243498.5 ENST00000379663.3 ENST00000509800.1 |
EXOSC9 |
exosome component 9 |
| chr7_+_80275953 | 0.06 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr14_-_71609965 | 0.06 |
ENST00000602957.1 |
RP6-91H8.3 |
RP6-91H8.3 |
| chr11_-_47447970 | 0.06 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr19_+_35773242 | 0.06 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr16_-_66764119 | 0.05 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr1_-_111970353 | 0.05 |
ENST00000369732.3 |
OVGP1 |
oviductal glycoprotein 1, 120kDa |
| chr4_+_25657444 | 0.05 |
ENST00000504570.1 ENST00000382051.3 |
SLC34A2 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr20_+_44421137 | 0.05 |
ENST00000415790.1 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr20_+_44420570 | 0.05 |
ENST00000372622.3 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr17_+_11501748 | 0.05 |
ENST00000262442.4 ENST00000579828.1 |
DNAH9 |
dynein, axonemal, heavy chain 9 |
| chr13_-_24895566 | 0.05 |
ENST00000422229.2 |
AL359736.1 |
protein PCOTH isoform 1 |
| chr7_+_23719749 | 0.05 |
ENST00000409192.3 ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A |
family with sequence similarity 221, member A |
| chr3_+_101498269 | 0.05 |
ENST00000491511.2 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr6_-_33041378 | 0.05 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr19_+_852291 | 0.05 |
ENST00000263621.1 |
ELANE |
elastase, neutrophil expressed |
| chr16_+_86612112 | 0.05 |
ENST00000320241.3 |
FOXL1 |
forkhead box L1 |
| chr14_+_72052983 | 0.05 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr11_+_120971882 | 0.05 |
ENST00000392793.1 |
TECTA |
tectorin alpha |
| chrX_-_41449204 | 0.05 |
ENST00000378179.3 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0097527 | Fas signaling pathway(GO:0036337) necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0002839 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 0.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 0.5 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0060230 | lipase binding(GO:0035473) lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |