Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for HOXA9
Z-value: 0.32
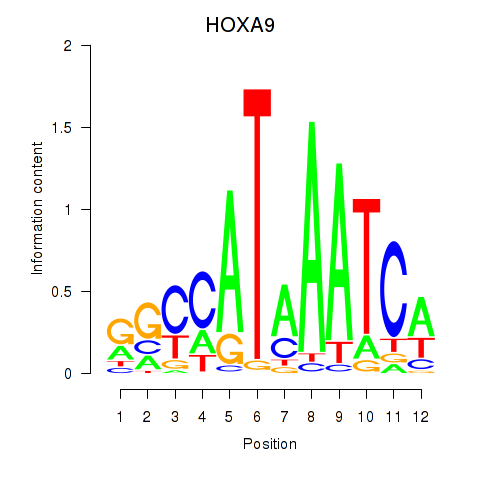
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | HOXA9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | 0.21 | 6.1e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_101702417 | 0.41 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr7_-_83824169 | 0.41 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_+_47666275 | 0.38 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr11_-_108464465 | 0.36 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr1_+_24645865 | 0.35 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.34 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr11_-_108464321 | 0.30 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr1_+_152956549 | 0.30 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr1_+_209602771 | 0.29 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr4_+_48018781 | 0.27 |
ENST00000295461.5 |
NIPAL1 |
NIPA-like domain containing 1 |
| chr17_-_56494908 | 0.27 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr7_+_69064300 | 0.27 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr9_-_27529726 | 0.26 |
ENST00000262244.5 |
MOB3B |
MOB kinase activator 3B |
| chr9_+_40028620 | 0.24 |
ENST00000426179.1 |
AL353791.1 |
AL353791.1 |
| chr7_+_20370746 | 0.23 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr5_+_147582387 | 0.19 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_24646002 | 0.19 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr18_+_34124507 | 0.19 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr4_-_74964904 | 0.19 |
ENST00000508487.2 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
| chr7_+_16793160 | 0.18 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr2_+_102608306 | 0.18 |
ENST00000332549.3 |
IL1R2 |
interleukin 1 receptor, type II |
| chr5_+_140213815 | 0.18 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr5_-_94417339 | 0.17 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr2_-_99870744 | 0.17 |
ENST00000409238.1 ENST00000423800.1 |
LYG2 |
lysozyme G-like 2 |
| chr8_+_31497271 | 0.16 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr6_-_51952367 | 0.16 |
ENST00000340994.4 |
PKHD1 |
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr10_+_24755416 | 0.16 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr4_-_77328458 | 0.16 |
ENST00000388914.3 ENST00000434846.2 |
CCDC158 |
coiled-coil domain containing 158 |
| chr17_-_7493390 | 0.16 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr6_+_25279651 | 0.15 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr1_-_85100703 | 0.15 |
ENST00000370624.1 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr5_-_54281491 | 0.15 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr1_-_24513737 | 0.15 |
ENST00000374421.3 ENST00000374418.3 ENST00000327535.1 ENST00000327575.2 |
IFNLR1 |
interferon, lambda receptor 1 |
| chr2_+_32502952 | 0.14 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr11_+_60048004 | 0.14 |
ENST00000532114.1 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr8_-_20040638 | 0.14 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr6_+_35310312 | 0.13 |
ENST00000448077.2 ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr4_-_186732048 | 0.13 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chrX_-_45629661 | 0.12 |
ENST00000602507.1 ENST00000602461.1 |
RP6-99M1.2 |
RP6-99M1.2 |
| chr11_-_57148619 | 0.12 |
ENST00000287143.2 |
PRG3 |
proteoglycan 3 |
| chr12_-_71148413 | 0.12 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr10_+_24738355 | 0.12 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr4_+_86749045 | 0.12 |
ENST00000514229.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_-_31637560 | 0.12 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr6_+_125304502 | 0.11 |
ENST00000519799.1 ENST00000368414.2 ENST00000359704.2 |
RNF217 |
ring finger protein 217 |
| chr7_-_92855762 | 0.11 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr10_-_45474237 | 0.11 |
ENST00000448778.1 ENST00000298295.3 |
C10orf10 |
chromosome 10 open reading frame 10 |
| chr5_+_147582348 | 0.11 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr9_+_21409146 | 0.11 |
ENST00000380205.1 |
IFNA8 |
interferon, alpha 8 |
| chr5_+_80529104 | 0.11 |
ENST00000254035.4 ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr3_+_97851542 | 0.11 |
ENST00000354565.2 |
OR5H1 |
olfactory receptor, family 5, subfamily H, member 1 |
| chr9_+_117373486 | 0.11 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr19_-_48547294 | 0.10 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chr5_+_126984710 | 0.10 |
ENST00000379445.3 |
CTXN3 |
cortexin 3 |
| chr2_+_143635067 | 0.10 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr5_-_43515231 | 0.10 |
ENST00000306862.2 |
C5orf34 |
chromosome 5 open reading frame 34 |
| chr12_-_10962767 | 0.10 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr16_-_20556492 | 0.10 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr7_-_3214287 | 0.10 |
ENST00000404626.3 |
AC091801.1 |
LOC392621; Uncharacterized protein |
| chr1_+_52082751 | 0.09 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr12_-_57472522 | 0.09 |
ENST00000379391.3 ENST00000300128.4 |
TMEM194A |
transmembrane protein 194A |
| chr11_-_66139617 | 0.09 |
ENST00000544554.1 ENST00000546034.1 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr14_-_106453155 | 0.09 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr11_-_59952106 | 0.09 |
ENST00000529054.1 ENST00000530839.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr2_+_143635222 | 0.09 |
ENST00000375773.2 ENST00000409512.1 ENST00000410015.2 |
KYNU |
kynureninase |
| chr11_-_66139570 | 0.09 |
ENST00000311161.7 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr9_-_127269661 | 0.09 |
ENST00000373588.4 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr6_-_9939552 | 0.09 |
ENST00000460363.2 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr11_+_34664014 | 0.09 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr1_-_11024258 | 0.09 |
ENST00000418570.2 |
C1orf127 |
chromosome 1 open reading frame 127 |
| chr2_+_128403439 | 0.09 |
ENST00000544369.1 |
GPR17 |
G protein-coupled receptor 17 |
| chr19_-_52598958 | 0.09 |
ENST00000594440.1 ENST00000426391.2 ENST00000389534.4 |
ZNF841 |
zinc finger protein 841 |
| chrX_+_65384182 | 0.09 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr2_-_209028300 | 0.09 |
ENST00000304502.4 |
CRYGA |
crystallin, gamma A |
| chr8_-_20040601 | 0.09 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr11_-_8959758 | 0.08 |
ENST00000531618.1 |
ASCL3 |
achaete-scute family bHLH transcription factor 3 |
| chr7_+_69064566 | 0.08 |
ENST00000403018.2 |
AUTS2 |
autism susceptibility candidate 2 |
| chr11_-_2162162 | 0.08 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr22_-_31364187 | 0.08 |
ENST00000215862.4 ENST00000397641.3 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr4_-_76944621 | 0.08 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr11_-_57092381 | 0.08 |
ENST00000358252.3 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
| chr7_+_55177416 | 0.08 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr1_+_44584522 | 0.08 |
ENST00000372299.3 |
KLF17 |
Kruppel-like factor 17 |
| chr14_+_23938891 | 0.08 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr12_-_121477039 | 0.08 |
ENST00000257570.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr6_+_35310391 | 0.08 |
ENST00000337400.2 ENST00000311565.4 ENST00000540939.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr8_+_110098850 | 0.08 |
ENST00000518632.1 |
TRHR |
thyrotropin-releasing hormone receptor |
| chr2_+_204732666 | 0.08 |
ENST00000295854.6 ENST00000472206.1 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr15_-_72668805 | 0.08 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr1_+_180897269 | 0.08 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr5_+_38845960 | 0.08 |
ENST00000502536.1 |
OSMR |
oncostatin M receptor |
| chr7_+_93535866 | 0.08 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr1_-_152552980 | 0.08 |
ENST00000368787.3 |
LCE3D |
late cornified envelope 3D |
| chr2_+_233390863 | 0.07 |
ENST00000449596.1 ENST00000543200.1 |
CHRND |
cholinergic receptor, nicotinic, delta (muscle) |
| chr7_+_93535817 | 0.07 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr5_-_101834617 | 0.07 |
ENST00000513675.1 ENST00000379807.3 |
SLCO6A1 |
solute carrier organic anion transporter family, member 6A1 |
| chr9_-_86571628 | 0.07 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr16_+_29690358 | 0.07 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr11_-_59951889 | 0.07 |
ENST00000532169.1 ENST00000534596.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr2_+_233390890 | 0.07 |
ENST00000258385.3 ENST00000536614.1 ENST00000457943.2 |
CHRND |
cholinergic receptor, nicotinic, delta (muscle) |
| chr2_-_130031335 | 0.07 |
ENST00000375987.3 |
AC079586.1 |
AC079586.1 |
| chr10_+_91092241 | 0.07 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr18_+_21032781 | 0.07 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr3_+_120626919 | 0.07 |
ENST00000273666.6 ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L |
syntaxin binding protein 5-like |
| chr2_+_171571827 | 0.07 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr22_+_32754139 | 0.07 |
ENST00000382088.3 |
RFPL3 |
ret finger protein-like 3 |
| chr4_+_86748898 | 0.06 |
ENST00000509300.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr11_+_19138670 | 0.06 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr3_-_72149553 | 0.06 |
ENST00000468646.2 ENST00000464271.1 |
LINC00877 |
long intergenic non-protein coding RNA 877 |
| chr18_-_72264805 | 0.06 |
ENST00000577806.1 |
LINC00909 |
long intergenic non-protein coding RNA 909 |
| chr11_+_6866883 | 0.06 |
ENST00000299454.4 ENST00000379831.2 |
OR10A5 |
olfactory receptor, family 10, subfamily A, member 5 |
| chr1_-_152297679 | 0.06 |
ENST00000368799.1 |
FLG |
filaggrin |
| chr9_-_21166659 | 0.06 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
| chr13_+_52586517 | 0.06 |
ENST00000523764.1 ENST00000521508.1 |
ALG11 |
ALG11, alpha-1,2-mannosyltransferase |
| chr14_-_74959978 | 0.06 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr15_-_54025300 | 0.06 |
ENST00000559418.1 |
WDR72 |
WD repeat domain 72 |
| chr3_-_58613323 | 0.06 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr4_-_72649763 | 0.06 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr4_-_100356551 | 0.06 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr18_+_42260861 | 0.06 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr7_-_16505440 | 0.06 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chr9_+_470288 | 0.06 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr14_-_74960030 | 0.06 |
ENST00000553490.1 ENST00000557510.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr9_-_95640218 | 0.06 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr4_+_113970772 | 0.06 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr5_-_37371163 | 0.06 |
ENST00000513532.1 |
NUP155 |
nucleoporin 155kDa |
| chr10_+_47894572 | 0.06 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr7_-_99332719 | 0.06 |
ENST00000336374.2 |
CYP3A7 |
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr12_+_25205666 | 0.06 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr6_-_25832254 | 0.06 |
ENST00000476801.1 ENST00000244527.4 ENST00000427328.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chr10_-_24770632 | 0.06 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr10_+_81967456 | 0.06 |
ENST00000422847.1 |
LINC00857 |
long intergenic non-protein coding RNA 857 |
| chr5_-_101834712 | 0.06 |
ENST00000506729.1 ENST00000389019.3 ENST00000379810.1 |
SLCO6A1 |
solute carrier organic anion transporter family, member 6A1 |
| chr5_+_34915444 | 0.06 |
ENST00000336767.5 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chrX_-_77150985 | 0.05 |
ENST00000358075.6 |
MAGT1 |
magnesium transporter 1 |
| chr6_-_137539651 | 0.05 |
ENST00000543628.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr17_-_39728303 | 0.05 |
ENST00000588431.1 ENST00000246662.4 |
KRT9 |
keratin 9 |
| chr14_-_74959994 | 0.05 |
ENST00000238633.2 ENST00000434013.2 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr14_-_65769392 | 0.05 |
ENST00000555736.1 |
CTD-2509G16.5 |
CTD-2509G16.5 |
| chr1_-_205819245 | 0.05 |
ENST00000367136.4 |
PM20D1 |
peptidase M20 domain containing 1 |
| chr6_+_18155560 | 0.05 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr12_-_13248598 | 0.05 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr2_+_238600933 | 0.05 |
ENST00000420665.1 ENST00000392000.4 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
| chr2_+_27255806 | 0.05 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chrX_+_114827818 | 0.05 |
ENST00000420625.2 |
PLS3 |
plastin 3 |
| chr17_+_60447579 | 0.05 |
ENST00000450662.2 |
EFCAB3 |
EF-hand calcium binding domain 3 |
| chr1_+_230193521 | 0.05 |
ENST00000543760.1 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr17_-_8113542 | 0.05 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr15_+_94899183 | 0.05 |
ENST00000557742.1 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
| chr1_-_216896780 | 0.05 |
ENST00000459955.1 ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_149230680 | 0.05 |
ENST00000443018.1 |
RP11-403I13.5 |
RP11-403I13.5 |
| chr19_-_6375860 | 0.05 |
ENST00000245810.1 |
PSPN |
persephin |
| chr5_+_38846101 | 0.05 |
ENST00000274276.3 |
OSMR |
oncostatin M receptor |
| chr3_-_143567262 | 0.05 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr7_+_21582638 | 0.05 |
ENST00000409508.3 ENST00000328843.6 |
DNAH11 |
dynein, axonemal, heavy chain 11 |
| chr3_+_108541608 | 0.05 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr5_-_37371278 | 0.05 |
ENST00000231498.3 |
NUP155 |
nucleoporin 155kDa |
| chr7_+_117251671 | 0.05 |
ENST00000468795.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr2_+_232063436 | 0.05 |
ENST00000440107.1 |
ARMC9 |
armadillo repeat containing 9 |
| chr4_-_100356291 | 0.05 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chrX_+_65384052 | 0.05 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr1_-_235292250 | 0.05 |
ENST00000366607.4 |
TOMM20 |
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr12_+_25205568 | 0.05 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr4_-_48018580 | 0.04 |
ENST00000514170.1 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr2_+_27799389 | 0.04 |
ENST00000408964.2 |
C2orf16 |
chromosome 2 open reading frame 16 |
| chr5_+_5422778 | 0.04 |
ENST00000296564.7 |
KIAA0947 |
KIAA0947 |
| chr17_-_48943706 | 0.04 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr7_-_144107320 | 0.04 |
ENST00000483238.1 ENST00000467773.1 |
NOBOX |
NOBOX oogenesis homeobox |
| chr4_-_71532339 | 0.04 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_113498616 | 0.04 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr3_+_119298429 | 0.04 |
ENST00000478927.1 |
ADPRH |
ADP-ribosylarginine hydrolase |
| chr1_+_166958346 | 0.04 |
ENST00000367872.4 |
MAEL |
maelstrom spermatogenic transposon silencer |
| chr12_-_121476959 | 0.04 |
ENST00000339275.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr16_-_18911366 | 0.04 |
ENST00000565224.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_+_118532937 | 0.04 |
ENST00000297347.3 |
MED30 |
mediator complex subunit 30 |
| chr11_+_85955787 | 0.04 |
ENST00000528180.1 |
EED |
embryonic ectoderm development |
| chr3_-_151176497 | 0.04 |
ENST00000282466.3 |
IGSF10 |
immunoglobulin superfamily, member 10 |
| chrX_+_130192318 | 0.04 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chrX_+_95939638 | 0.04 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr15_-_68497657 | 0.04 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr6_-_46138676 | 0.04 |
ENST00000371383.2 ENST00000230565.3 |
ENPP5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr4_-_102267953 | 0.04 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_-_133035185 | 0.04 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr3_-_108672742 | 0.04 |
ENST00000261047.3 |
GUCA1C |
guanylate cyclase activator 1C |
| chrX_+_30248553 | 0.04 |
ENST00000361644.2 |
MAGEB3 |
melanoma antigen family B, 3 |
| chr15_+_65134088 | 0.04 |
ENST00000323544.4 ENST00000437723.1 |
PLEKHO2 AC069368.3 |
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr16_-_55867146 | 0.04 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr11_+_61891445 | 0.04 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr12_+_122181529 | 0.04 |
ENST00000541467.1 |
TMEM120B |
transmembrane protein 120B |
| chr11_-_104893863 | 0.04 |
ENST00000260315.3 ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5 |
caspase 5, apoptosis-related cysteine peptidase |
| chr6_+_130339710 | 0.04 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr3_-_108672609 | 0.04 |
ENST00000393963.3 ENST00000471108.1 |
GUCA1C |
guanylate cyclase activator 1C |
| chr15_-_31283618 | 0.04 |
ENST00000563714.1 |
MTMR10 |
myotubularin related protein 10 |
| chr6_+_160327974 | 0.04 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr12_-_13248732 | 0.04 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr18_+_21033239 | 0.04 |
ENST00000581585.1 ENST00000577501.1 |
RIOK3 |
RIO kinase 3 |
| chr10_+_5488564 | 0.04 |
ENST00000449083.1 ENST00000380359.3 |
NET1 |
neuroepithelial cell transforming 1 |
| chr4_+_70796784 | 0.04 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr15_-_41166414 | 0.04 |
ENST00000220507.4 |
RHOV |
ras homolog family member V |
| chr11_-_2182388 | 0.04 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.4 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.4 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.9 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.0 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.0 | 0.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |