Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
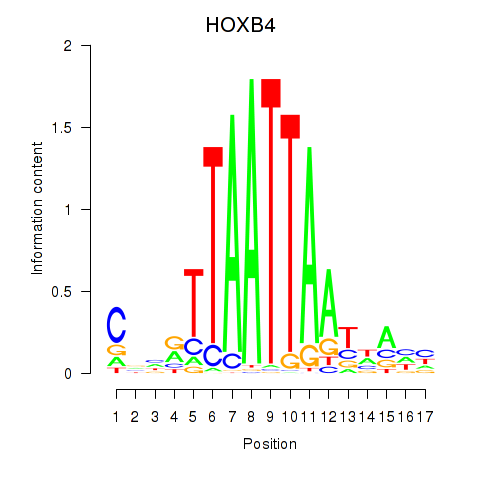
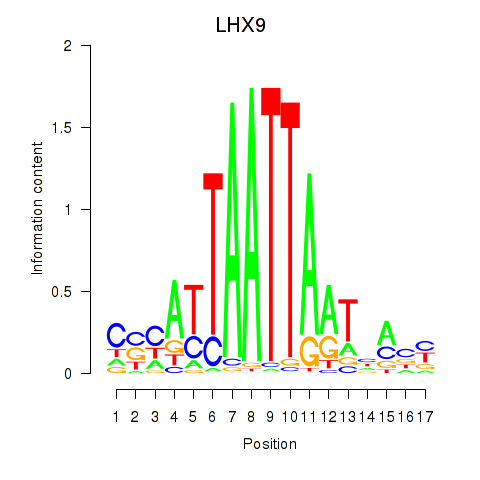
Results for HOXB4_LHX9
Z-value: 1.21
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB4
|
ENSG00000182742.5 | HOXB4 |
|
LHX9
|
ENSG00000143355.11 | LHX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB4 | hg19_v2_chr17_-_46657473_46657473 | 0.92 | 1.0e-03 | Click! |
| LHX9 | hg19_v2_chr1_+_197881592_197881635 | -0.57 | 1.4e-01 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_105845674 | 2.38 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr19_-_36001113 | 1.82 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr12_-_8803128 | 1.61 |
ENST00000543467.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr2_-_161056762 | 1.40 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056802 | 1.39 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr18_+_29027696 | 1.29 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr1_+_160370344 | 1.25 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr8_-_41166953 | 1.08 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr1_+_62439037 | 1.07 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr5_+_66300446 | 0.90 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_28261492 | 0.85 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr3_-_151034734 | 0.74 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr18_+_21452964 | 0.74 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr5_+_89770664 | 0.73 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr18_+_21452804 | 0.72 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr5_+_89770696 | 0.72 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr10_+_24755416 | 0.71 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr2_+_182850551 | 0.65 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_-_5323226 | 0.62 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr11_-_119991589 | 0.62 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr10_+_6779326 | 0.59 |
ENST00000417112.1 |
RP11-554I8.2 |
RP11-554I8.2 |
| chr6_+_130339710 | 0.57 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr17_-_39743139 | 0.57 |
ENST00000167586.6 |
KRT14 |
keratin 14 |
| chr14_-_67878917 | 0.57 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr16_-_28634874 | 0.55 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr18_-_74839891 | 0.53 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr2_+_182850743 | 0.51 |
ENST00000409702.1 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_242612779 | 0.50 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr7_-_25702669 | 0.48 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr4_-_74486109 | 0.45 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_111718036 | 0.44 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr8_+_105235572 | 0.44 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr3_+_111717600 | 0.43 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_-_74486217 | 0.43 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_+_124329336 | 0.41 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr3_+_111718173 | 0.41 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr3_+_111717511 | 0.39 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr8_-_125577940 | 0.39 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chr4_+_76481258 | 0.39 |
ENST00000311623.4 ENST00000435974.2 |
C4orf26 |
chromosome 4 open reading frame 26 |
| chr2_+_102615416 | 0.39 |
ENST00000393414.2 |
IL1R2 |
interleukin 1 receptor, type II |
| chr5_-_35938674 | 0.39 |
ENST00000397366.1 ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL |
calcyphosine-like |
| chr4_-_159956333 | 0.38 |
ENST00000434826.2 |
C4orf45 |
chromosome 4 open reading frame 45 |
| chr3_-_141747950 | 0.37 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_90211643 | 0.37 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr5_+_140220769 | 0.35 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr17_-_19015945 | 0.34 |
ENST00000573866.2 |
SNORD3D |
small nucleolar RNA, C/D box 3D |
| chr6_-_32806506 | 0.33 |
ENST00000374897.2 ENST00000452392.2 |
TAP2 TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr4_-_90757364 | 0.33 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_152784447 | 0.33 |
ENST00000360090.3 |
LCE1B |
late cornified envelope 1B |
| chr19_-_36304201 | 0.30 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr20_+_31805131 | 0.30 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr11_-_128894053 | 0.29 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr6_+_26365443 | 0.29 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr16_-_30122717 | 0.28 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_90273679 | 0.28 |
ENST00000423080.2 |
IGKV3D-7 |
immunoglobulin kappa variable 3D-7 |
| chrX_-_100662881 | 0.27 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr14_+_22670455 | 0.27 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr20_-_29978383 | 0.26 |
ENST00000339144.3 ENST00000376321.3 |
DEFB119 |
defensin, beta 119 |
| chr14_+_103851712 | 0.26 |
ENST00000440884.3 ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
| chr11_-_104827425 | 0.26 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_59522837 | 0.25 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr6_+_42584847 | 0.25 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr21_-_31538971 | 0.25 |
ENST00000286808.3 |
CLDN17 |
claudin 17 |
| chr17_+_19091325 | 0.24 |
ENST00000584923.1 |
SNORD3A |
small nucleolar RNA, C/D box 3A |
| chr11_-_107729887 | 0.24 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr11_-_75917569 | 0.24 |
ENST00000322563.3 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
| chr12_-_52604607 | 0.23 |
ENST00000551894.1 ENST00000553017.1 |
C12orf80 |
chromosome 12 open reading frame 80 |
| chr11_+_55594695 | 0.23 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr7_+_115862858 | 0.23 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr4_+_119810134 | 0.22 |
ENST00000434046.2 |
SYNPO2 |
synaptopodin 2 |
| chr4_-_74486347 | 0.22 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_+_196743912 | 0.22 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr2_-_130031335 | 0.22 |
ENST00000375987.3 |
AC079586.1 |
AC079586.1 |
| chr3_-_191000172 | 0.22 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr2_-_89327228 | 0.21 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr12_-_112279694 | 0.21 |
ENST00000443596.1 ENST00000442119.1 |
MAPKAPK5-AS1 |
MAPKAPK5 antisense RNA 1 |
| chr20_-_50722183 | 0.21 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr7_-_111032971 | 0.20 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr21_-_42219065 | 0.20 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chrX_+_37639302 | 0.20 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr7_-_37024665 | 0.20 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr4_+_110749143 | 0.19 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_+_81110684 | 0.19 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr11_-_46408107 | 0.19 |
ENST00000433765.2 |
CHRM4 |
cholinergic receptor, muscarinic 4 |
| chr3_-_185538849 | 0.19 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr17_-_38821373 | 0.18 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr3_-_141719195 | 0.18 |
ENST00000397991.4 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr19_+_11485333 | 0.18 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr14_+_31046959 | 0.17 |
ENST00000547532.1 ENST00000555429.1 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
| chr6_+_26402465 | 0.17 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr14_-_72458326 | 0.17 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr14_+_24641062 | 0.17 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr2_+_234826016 | 0.17 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_+_90077680 | 0.17 |
ENST00000390270.2 |
IGKV3D-20 |
immunoglobulin kappa variable 3D-20 |
| chr1_+_81771806 | 0.16 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr12_-_91451758 | 0.16 |
ENST00000266719.3 |
KERA |
keratocan |
| chr15_-_72523924 | 0.16 |
ENST00000566809.1 ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM |
pyruvate kinase, muscle |
| chr6_+_129204337 | 0.16 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr2_+_234637754 | 0.16 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_102456277 | 0.16 |
ENST00000421882.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_-_120243545 | 0.16 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr6_+_26402517 | 0.16 |
ENST00000414912.2 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr14_-_104181771 | 0.15 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr12_-_22063787 | 0.15 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr6_-_29527702 | 0.15 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr4_-_39979576 | 0.15 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr10_+_695888 | 0.14 |
ENST00000441152.2 |
PRR26 |
proline rich 26 |
| chr6_+_3259148 | 0.14 |
ENST00000419065.2 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4 |
proteasome (prosome, macropain) assembly chaperone 4 |
| chr2_+_102953608 | 0.14 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr8_-_33424636 | 0.14 |
ENST00000256257.1 |
RNF122 |
ring finger protein 122 |
| chr1_-_67266939 | 0.14 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr13_+_32313658 | 0.14 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr4_-_103997862 | 0.14 |
ENST00000394785.3 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr4_-_103998439 | 0.14 |
ENST00000503230.1 ENST00000503818.1 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr2_+_202047596 | 0.14 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr4_+_156587853 | 0.13 |
ENST00000506455.1 ENST00000511108.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr7_-_87856280 | 0.13 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr5_+_36152179 | 0.13 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr10_+_127585093 | 0.13 |
ENST00000368695.1 ENST00000368693.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr15_-_82641706 | 0.13 |
ENST00000439287.4 |
GOLGA6L10 |
golgin A6 family-like 10 |
| chr7_-_87856303 | 0.13 |
ENST00000394641.3 |
SRI |
sorcin |
| chr12_+_20963632 | 0.13 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr3_-_20053741 | 0.13 |
ENST00000389050.4 |
PP2D1 |
protein phosphatase 2C-like domain containing 1 |
| chr9_-_77643307 | 0.13 |
ENST00000376834.3 ENST00000376830.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr10_+_101491968 | 0.13 |
ENST00000370476.5 ENST00000370472.4 |
CUTC |
cutC copper transporter |
| chrX_-_71458802 | 0.13 |
ENST00000373657.1 ENST00000334463.3 |
ERCC6L |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr2_-_128284020 | 0.12 |
ENST00000295321.4 ENST00000455721.2 |
IWS1 |
IWS1 homolog (S. cerevisiae) |
| chr11_+_59824127 | 0.12 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chrX_+_100663243 | 0.12 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr4_+_100432161 | 0.12 |
ENST00000326581.4 ENST00000514652.1 |
C4orf17 |
chromosome 4 open reading frame 17 |
| chr3_-_160823040 | 0.12 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr12_+_10163231 | 0.12 |
ENST00000396502.1 ENST00000338896.5 |
CLEC12B |
C-type lectin domain family 12, member B |
| chr8_+_50824233 | 0.12 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr4_+_86525299 | 0.12 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr13_+_97928395 | 0.12 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr12_+_112279782 | 0.12 |
ENST00000550735.2 |
MAPKAPK5 |
mitogen-activated protein kinase-activated protein kinase 5 |
| chr3_-_160823158 | 0.12 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr17_-_4938712 | 0.12 |
ENST00000254853.5 ENST00000424747.1 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
| chr2_+_202047843 | 0.12 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr2_-_43266680 | 0.12 |
ENST00000425212.1 ENST00000422351.1 ENST00000449766.1 |
AC016735.2 |
AC016735.2 |
| chr5_+_36152091 | 0.12 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr4_+_108745711 | 0.12 |
ENST00000394684.4 |
SGMS2 |
sphingomyelin synthase 2 |
| chr9_-_95640218 | 0.11 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr15_+_66679155 | 0.11 |
ENST00000307102.5 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
| chr8_-_101571964 | 0.11 |
ENST00000520552.1 ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr8_+_107738343 | 0.11 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr17_+_18086392 | 0.11 |
ENST00000541285.1 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
| chr16_-_28608364 | 0.11 |
ENST00000533150.1 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr18_-_51751132 | 0.11 |
ENST00000256429.3 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr1_+_161691353 | 0.11 |
ENST00000367948.2 |
FCRLB |
Fc receptor-like B |
| chr9_+_117904097 | 0.11 |
ENST00000374016.1 |
DEC1 |
deleted in esophageal cancer 1 |
| chr2_-_169887827 | 0.11 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr5_+_36152163 | 0.11 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_-_89385283 | 0.11 |
ENST00000390252.2 |
IGKV3-15 |
immunoglobulin kappa variable 3-15 |
| chr11_+_7626950 | 0.11 |
ENST00000530181.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr16_+_15489603 | 0.11 |
ENST00000568766.1 ENST00000287594.7 |
RP11-1021N1.1 MPV17L |
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr11_+_59824060 | 0.11 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr5_+_55149150 | 0.10 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr1_-_201140673 | 0.10 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr14_+_56584414 | 0.10 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr6_+_30585486 | 0.10 |
ENST00000259873.4 ENST00000506373.2 |
MRPS18B |
mitochondrial ribosomal protein S18B |
| chr21_-_15583165 | 0.10 |
ENST00000536861.1 |
LIPI |
lipase, member I |
| chr20_+_30697298 | 0.10 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr8_-_101571933 | 0.10 |
ENST00000520311.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr4_+_80584903 | 0.10 |
ENST00000506460.1 |
RP11-452C8.1 |
RP11-452C8.1 |
| chr12_+_20963647 | 0.10 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_170948361 | 0.10 |
ENST00000393702.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr2_+_87565634 | 0.10 |
ENST00000421835.2 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr14_+_23938891 | 0.10 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr11_+_118398178 | 0.10 |
ENST00000302783.4 ENST00000539546.1 |
TTC36 |
tetratricopeptide repeat domain 36 |
| chr2_-_90538397 | 0.10 |
ENST00000443397.3 |
RP11-685N3.1 |
Uncharacterized protein |
| chr9_+_125132803 | 0.10 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_1225470 | 0.10 |
ENST00000324642.3 ENST00000296821.4 |
SLC6A18 |
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr19_-_3557570 | 0.09 |
ENST00000355415.2 |
MFSD12 |
major facilitator superfamily domain containing 12 |
| chr13_+_48611665 | 0.09 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr12_-_112123524 | 0.09 |
ENST00000327551.6 |
BRAP |
BRCA1 associated protein |
| chr1_-_39339777 | 0.09 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr7_-_44580861 | 0.09 |
ENST00000546276.1 ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1 |
NPC1-like 1 |
| chr4_-_103998060 | 0.09 |
ENST00000339611.4 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr7_-_44122063 | 0.09 |
ENST00000335195.6 ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM |
polymerase (DNA directed), mu |
| chr1_-_205091115 | 0.09 |
ENST00000264515.6 ENST00000367164.1 |
RBBP5 |
retinoblastoma binding protein 5 |
| chr3_-_194072019 | 0.09 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr3_+_139063372 | 0.09 |
ENST00000478464.1 |
MRPS22 |
mitochondrial ribosomal protein S22 |
| chr3_+_23851928 | 0.09 |
ENST00000467766.1 ENST00000424381.1 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr15_+_58702742 | 0.08 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr16_-_29934558 | 0.08 |
ENST00000568995.1 ENST00000566413.1 |
KCTD13 |
potassium channel tetramerization domain containing 13 |
| chr7_+_107224364 | 0.08 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr6_+_29079668 | 0.08 |
ENST00000377169.1 |
OR2J3 |
olfactory receptor, family 2, subfamily J, member 3 |
| chr7_+_150020329 | 0.08 |
ENST00000323078.7 |
LRRC61 |
leucine rich repeat containing 61 |
| chr15_-_55489097 | 0.08 |
ENST00000260443.4 |
RSL24D1 |
ribosomal L24 domain containing 1 |
| chr14_-_106552755 | 0.08 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr5_+_140557371 | 0.08 |
ENST00000239444.2 |
PCDHB8 |
protocadherin beta 8 |
| chr2_-_101925055 | 0.08 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr1_-_197115818 | 0.08 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr7_-_81399411 | 0.08 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_+_1710484 | 0.08 |
ENST00000313871.3 ENST00000381261.3 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
| chr10_+_90424196 | 0.08 |
ENST00000394375.3 ENST00000608620.1 ENST00000238983.4 ENST00000355843.2 |
LIPF |
lipase, gastric |
| chr1_-_92952433 | 0.08 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr2_-_136678123 | 0.07 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr11_+_122753391 | 0.07 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.4 | 1.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.3 | 1.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 2.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.2 | 4.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.4 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.4 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 1.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.0 | GO:0070640 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0036030 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.4 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 2.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |