Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
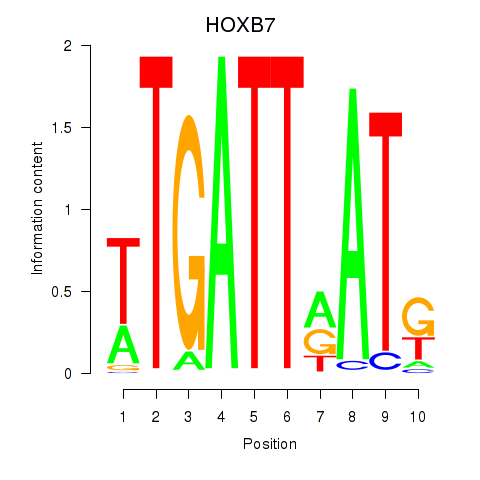
Results for HOXB7
Z-value: 0.83
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | HOXB7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | -0.11 | 7.9e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 1.71 |
ENST00000550758.1 |
DCN |
decorin |
| chr12_-_91573132 | 1.41 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr4_-_70626314 | 1.13 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr10_+_69865866 | 1.12 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr4_-_70626430 | 0.99 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr3_+_141105705 | 0.91 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr2_-_188378368 | 0.87 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr10_+_124320195 | 0.85 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr10_+_124320156 | 0.84 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr2_-_190044480 | 0.77 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr12_-_91573316 | 0.76 |
ENST00000393155.1 |
DCN |
decorin |
| chr12_+_59989918 | 0.73 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_+_59989791 | 0.72 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_9431324 | 0.72 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr11_-_89223883 | 0.72 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_+_45927994 | 0.71 |
ENST00000357632.2 ENST00000395963.2 |
CCR9 |
chemokine (C-C motif) receptor 9 |
| chr13_-_33760216 | 0.64 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr7_-_138363824 | 0.60 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr4_-_118006697 | 0.57 |
ENST00000310754.4 |
TRAM1L1 |
translocation associated membrane protein 1-like 1 |
| chr7_+_120629653 | 0.53 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_15596123 | 0.52 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr3_+_141105235 | 0.50 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_-_46507567 | 0.49 |
ENST00000584924.1 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr10_-_106240032 | 0.48 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr14_+_101299520 | 0.47 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr2_-_145278475 | 0.45 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr5_-_75919253 | 0.44 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr14_+_51955831 | 0.44 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr1_-_85100703 | 0.41 |
ENST00000370624.1 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr11_-_27723158 | 0.41 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr12_-_91573249 | 0.41 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr19_+_50016610 | 0.40 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_63376013 | 0.39 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr2_-_200322723 | 0.38 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr9_+_40028620 | 0.37 |
ENST00000426179.1 |
AL353791.1 |
AL353791.1 |
| chr5_+_98109322 | 0.37 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr20_+_5931497 | 0.35 |
ENST00000378886.2 ENST00000265187.4 |
MCM8 |
minichromosome maintenance complex component 8 |
| chr3_+_141106458 | 0.35 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr7_-_47579188 | 0.35 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr18_-_10748498 | 0.34 |
ENST00000579949.1 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr13_-_110438914 | 0.31 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr6_-_51952367 | 0.31 |
ENST00000340994.4 |
PKHD1 |
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr3_+_148415571 | 0.30 |
ENST00000497524.1 ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr12_+_52056548 | 0.30 |
ENST00000545061.1 ENST00000355133.3 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
| chr10_+_47894572 | 0.30 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr19_+_55315087 | 0.28 |
ENST00000345540.5 ENST00000357494.4 ENST00000396293.1 ENST00000346587.4 ENST00000396289.5 |
KIR2DL4 |
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr1_-_241520525 | 0.28 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr2_+_20646824 | 0.28 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr6_+_144904334 | 0.27 |
ENST00000367526.4 |
UTRN |
utrophin |
| chr1_-_167059830 | 0.27 |
ENST00000367868.3 |
GPA33 |
glycoprotein A33 (transmembrane) |
| chr11_-_2162162 | 0.26 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr14_-_74417096 | 0.26 |
ENST00000286544.3 |
FAM161B |
family with sequence similarity 161, member B |
| chr19_-_20748541 | 0.25 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr16_-_46655538 | 0.25 |
ENST00000303383.3 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
| chr3_-_145878954 | 0.25 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr16_+_22501658 | 0.25 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr15_-_37393406 | 0.25 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr6_+_114178512 | 0.24 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr19_-_52598958 | 0.24 |
ENST00000594440.1 ENST00000426391.2 ENST00000389534.4 |
ZNF841 |
zinc finger protein 841 |
| chr10_+_47894023 | 0.24 |
ENST00000358474.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr8_-_108510224 | 0.24 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr5_-_38557561 | 0.23 |
ENST00000511561.1 |
LIFR |
leukemia inhibitory factor receptor alpha |
| chr6_+_47624172 | 0.23 |
ENST00000507065.1 ENST00000296862.1 |
GPR111 |
G protein-coupled receptor 111 |
| chr1_-_146068184 | 0.23 |
ENST00000604894.1 ENST00000369323.3 ENST00000479926.2 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr3_+_141106643 | 0.23 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr7_-_27142290 | 0.22 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chrX_+_57618269 | 0.22 |
ENST00000374888.1 |
ZXDB |
zinc finger, X-linked, duplicated B |
| chrX_-_92928557 | 0.22 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr10_+_17270214 | 0.22 |
ENST00000544301.1 |
VIM |
vimentin |
| chr14_+_55034599 | 0.22 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr16_+_50099852 | 0.22 |
ENST00000299192.7 ENST00000285767.4 |
HEATR3 |
HEAT repeat containing 3 |
| chr11_-_70507901 | 0.22 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_117043 | 0.22 |
ENST00000320901.3 |
OR4F21 |
olfactory receptor, family 4, subfamily F, member 21 |
| chr1_-_147610081 | 0.22 |
ENST00000369226.3 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr3_+_111393501 | 0.22 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chrX_+_36053908 | 0.21 |
ENST00000378660.2 |
CHDC2 |
calponin homology domain containing 2 |
| chr20_+_36531499 | 0.21 |
ENST00000373458.3 ENST00000373461.4 ENST00000373459.4 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
| chr5_+_140772381 | 0.21 |
ENST00000398604.2 |
PCDHGA8 |
protocadherin gamma subfamily A, 8 |
| chr3_-_151102529 | 0.21 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr1_+_12524965 | 0.21 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_+_191046810 | 0.21 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr3_+_148545586 | 0.21 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr6_-_13621126 | 0.20 |
ENST00000600057.1 |
AL441883.1 |
Uncharacterized protein |
| chr11_-_5345582 | 0.20 |
ENST00000328813.2 |
OR51B2 |
olfactory receptor, family 51, subfamily B, member 2 |
| chr10_+_13142075 | 0.20 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr16_-_88729473 | 0.20 |
ENST00000301012.3 ENST00000569177.1 |
MVD |
mevalonate (diphospho) decarboxylase |
| chr5_-_10761206 | 0.20 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr6_+_167704838 | 0.19 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr11_+_6947647 | 0.19 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr2_+_63277927 | 0.19 |
ENST00000282549.2 |
OTX1 |
orthodenticle homeobox 1 |
| chr11_-_57194948 | 0.19 |
ENST00000533235.1 ENST00000526621.1 ENST00000352187.1 |
SLC43A3 |
solute carrier family 43, member 3 |
| chr11_-_96123022 | 0.19 |
ENST00000542662.1 |
CCDC82 |
coiled-coil domain containing 82 |
| chr10_+_13142225 | 0.19 |
ENST00000378747.3 |
OPTN |
optineurin |
| chrX_+_135067576 | 0.19 |
ENST00000370701.1 ENST00000370698.3 ENST00000370695.4 |
SLC9A6 |
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
| chr18_-_77276057 | 0.19 |
ENST00000597412.1 |
AC018445.1 |
Uncharacterized protein |
| chr9_-_73736511 | 0.19 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_-_112106578 | 0.18 |
ENST00000369717.4 |
ADORA3 |
adenosine A3 receptor |
| chr14_+_62229075 | 0.18 |
ENST00000216294.4 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr10_-_65028938 | 0.18 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr10_+_48189612 | 0.18 |
ENST00000453919.1 |
AGAP9 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr4_-_177116772 | 0.18 |
ENST00000280191.2 |
SPATA4 |
spermatogenesis associated 4 |
| chr5_-_75919217 | 0.18 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr5_+_54455946 | 0.18 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr1_-_165414414 | 0.18 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chrM_+_9207 | 0.18 |
ENST00000362079.2 |
MT-CO3 |
mitochondrially encoded cytochrome c oxidase III |
| chr1_+_367640 | 0.17 |
ENST00000426406.1 |
OR4F29 |
olfactory receptor, family 4, subfamily F, member 29 |
| chr3_-_93692781 | 0.17 |
ENST00000394236.3 |
PROS1 |
protein S (alpha) |
| chr6_+_45390222 | 0.17 |
ENST00000359524.5 |
RUNX2 |
runt-related transcription factor 2 |
| chr6_+_34204642 | 0.17 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr19_+_55315059 | 0.17 |
ENST00000359085.4 |
KIR2DL4 |
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr15_+_84904525 | 0.17 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr2_-_27886676 | 0.16 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr1_-_104238912 | 0.16 |
ENST00000330330.5 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr12_-_7596735 | 0.16 |
ENST00000416109.2 ENST00000396630.1 ENST00000313599.3 |
CD163L1 |
CD163 molecule-like 1 |
| chrX_-_39923656 | 0.16 |
ENST00000413905.1 |
BCOR |
BCL6 corepressor |
| chr3_+_108321623 | 0.16 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr5_-_171881362 | 0.16 |
ENST00000519643.1 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr15_+_85923856 | 0.16 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr1_+_224370873 | 0.15 |
ENST00000323699.4 ENST00000391877.3 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
| chr16_-_18462221 | 0.15 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr15_+_82722225 | 0.15 |
ENST00000300515.8 |
GOLGA6L9 |
golgin A6 family-like 9 |
| chr16_-_51185172 | 0.15 |
ENST00000251020.4 |
SALL1 |
spalt-like transcription factor 1 |
| chr12_+_98909351 | 0.15 |
ENST00000343315.5 ENST00000266732.4 ENST00000393053.2 |
TMPO |
thymopoietin |
| chr19_+_44645700 | 0.15 |
ENST00000592437.1 |
ZNF234 |
zinc finger protein 234 |
| chr1_+_104198377 | 0.15 |
ENST00000370083.4 |
AMY1A |
amylase, alpha 1A (salivary) |
| chr7_+_75024903 | 0.15 |
ENST00000323819.3 ENST00000430211.1 |
TRIM73 |
tripartite motif containing 73 |
| chr3_+_159943362 | 0.15 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chrX_-_106243451 | 0.15 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr7_+_116654935 | 0.15 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr3_+_189507460 | 0.15 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr12_-_65146636 | 0.15 |
ENST00000418919.2 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
| chr1_-_104239076 | 0.15 |
ENST00000370080.3 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr15_+_64680003 | 0.15 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr5_-_138534071 | 0.15 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr8_-_116681221 | 0.15 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr15_+_85923797 | 0.15 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr10_-_47239738 | 0.15 |
ENST00000413193.2 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr12_+_21679220 | 0.15 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr5_+_162887556 | 0.14 |
ENST00000393915.4 ENST00000432118.2 ENST00000358715.3 |
HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
| chrX_+_95939638 | 0.14 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr7_-_120498357 | 0.14 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr5_+_57787254 | 0.14 |
ENST00000502276.1 ENST00000396776.2 ENST00000511930.1 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
| chr16_+_66442411 | 0.14 |
ENST00000499966.1 |
LINC00920 |
long intergenic non-protein coding RNA 920 |
| chr6_+_72926145 | 0.14 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr16_-_66584059 | 0.14 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr6_-_44281043 | 0.14 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr11_-_111649015 | 0.14 |
ENST00000529841.1 |
RP11-108O10.2 |
RP11-108O10.2 |
| chr19_-_51920952 | 0.14 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr19_+_20959142 | 0.14 |
ENST00000344519.8 |
ZNF66 |
zinc finger protein 66 |
| chr2_-_154335300 | 0.14 |
ENST00000325926.3 |
RPRM |
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr9_-_85882145 | 0.14 |
ENST00000328788.1 |
FRMD3 |
FERM domain containing 3 |
| chrX_+_88002226 | 0.14 |
ENST00000276127.4 ENST00000373111.1 |
CPXCR1 |
CPX chromosome region, candidate 1 |
| chr1_-_622053 | 0.14 |
ENST00000332831.2 |
OR4F16 |
olfactory receptor, family 4, subfamily F, member 16 |
| chr17_+_41363854 | 0.13 |
ENST00000588693.1 ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A |
transmembrane protein 106A |
| chr7_-_86595190 | 0.13 |
ENST00000398276.2 ENST00000416314.1 ENST00000425689.1 |
KIAA1324L |
KIAA1324-like |
| chr15_+_23255242 | 0.13 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr17_-_7218631 | 0.13 |
ENST00000577040.2 ENST00000389167.5 ENST00000391950.3 |
GPS2 |
G protein pathway suppressor 2 |
| chr2_+_44396000 | 0.13 |
ENST00000409895.4 ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr10_-_17243579 | 0.13 |
ENST00000525762.1 ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
| chr8_+_56685701 | 0.13 |
ENST00000260129.5 |
TGS1 |
trimethylguanosine synthase 1 |
| chr8_-_116680833 | 0.13 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr18_+_29171689 | 0.13 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr16_-_66583701 | 0.13 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr10_+_104535994 | 0.13 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr2_-_27886460 | 0.13 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr10_-_14996321 | 0.13 |
ENST00000378289.4 |
DCLRE1C |
DNA cross-link repair 1C |
| chr17_-_39203519 | 0.13 |
ENST00000542137.1 ENST00000391419.3 |
KRTAP2-1 |
keratin associated protein 2-1 |
| chr15_-_37390482 | 0.12 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr5_+_180650271 | 0.12 |
ENST00000351937.5 ENST00000315073.5 |
TRIM41 |
tripartite motif containing 41 |
| chr8_+_41386761 | 0.12 |
ENST00000523277.2 |
GINS4 |
GINS complex subunit 4 (Sld5 homolog) |
| chr11_+_96123158 | 0.12 |
ENST00000332349.4 ENST00000458427.1 |
JRKL |
jerky homolog-like (mouse) |
| chr2_+_27886330 | 0.12 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr16_+_86612112 | 0.12 |
ENST00000320241.3 |
FOXL1 |
forkhead box L1 |
| chr3_-_108836977 | 0.12 |
ENST00000232603.5 |
MORC1 |
MORC family CW-type zinc finger 1 |
| chrX_+_130192318 | 0.12 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr1_-_179112173 | 0.12 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr9_+_2029019 | 0.12 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_-_93616340 | 0.12 |
ENST00000557420.1 ENST00000542321.2 |
RGMA |
repulsive guidance molecule family member a |
| chr11_+_844406 | 0.12 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr1_-_200589859 | 0.11 |
ENST00000367350.4 |
KIF14 |
kinesin family member 14 |
| chr15_+_83098710 | 0.11 |
ENST00000561062.1 ENST00000358583.3 |
GOLGA6L9 |
golgin A6 family-like 20 |
| chr17_-_8770956 | 0.11 |
ENST00000311434.9 |
PIK3R6 |
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr12_+_56401268 | 0.11 |
ENST00000262032.5 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
| chr11_+_117198545 | 0.11 |
ENST00000533153.1 ENST00000278935.3 ENST00000525416.1 |
CEP164 |
centrosomal protein 164kDa |
| chr4_+_144106080 | 0.11 |
ENST00000307017.4 |
USP38 |
ubiquitin specific peptidase 38 |
| chr8_+_42128812 | 0.11 |
ENST00000520810.1 ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr3_-_42452050 | 0.11 |
ENST00000441172.1 ENST00000287748.3 |
LYZL4 |
lysozyme-like 4 |
| chr6_+_89674246 | 0.11 |
ENST00000369474.1 |
AL079342.1 |
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr16_-_49890016 | 0.11 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr9_+_104161123 | 0.11 |
ENST00000374861.3 ENST00000339664.2 ENST00000259395.4 |
ZNF189 |
zinc finger protein 189 |
| chr9_+_124103625 | 0.11 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr4_+_158493642 | 0.11 |
ENST00000507108.1 ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1 |
RP11-364P22.1 |
| chr8_+_28748765 | 0.11 |
ENST00000355231.5 |
HMBOX1 |
homeobox containing 1 |
| chr14_+_50065376 | 0.11 |
ENST00000298288.6 |
LRR1 |
leucine rich repeat protein 1 |
| chr14_-_94789663 | 0.11 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr14_+_56078695 | 0.10 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr2_-_161350305 | 0.10 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr12_-_102455902 | 0.10 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr15_+_28623784 | 0.10 |
ENST00000526619.2 ENST00000337838.7 ENST00000532622.2 |
GOLGA8F |
golgin A8 family, member F |
| chr4_+_95128748 | 0.10 |
ENST00000359052.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.5 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 1.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 2.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0070837 | L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 1.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.2 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 1.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 4.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 2.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0033300 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) dehydroascorbic acid transporter activity(GO:0033300) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |