Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
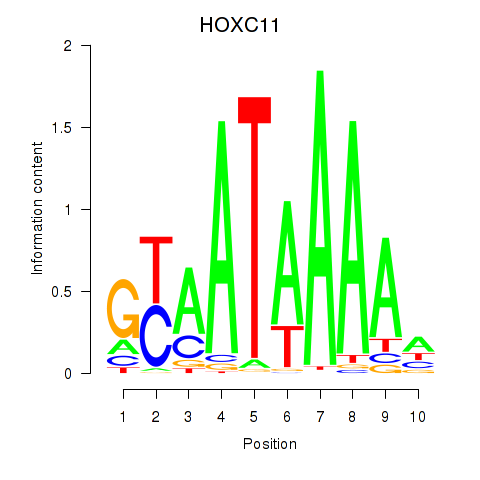
Results for HOXC11
Z-value: 0.78
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | HOXC11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | 0.70 | 5.4e-02 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_58179582 | 0.97 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr8_+_104831472 | 0.68 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr1_+_62439037 | 0.56 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_-_28124903 | 0.54 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr9_-_124989804 | 0.51 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr20_+_52105495 | 0.51 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr7_-_41742697 | 0.44 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr12_-_52828147 | 0.41 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr17_-_39646116 | 0.38 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr8_+_104831554 | 0.37 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr18_-_52989217 | 0.37 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr16_+_57653989 | 0.35 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57653854 | 0.35 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_-_12677714 | 0.33 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chrX_-_133792480 | 0.32 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr9_-_124990680 | 0.31 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr5_-_82969405 | 0.30 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr9_+_12693336 | 0.29 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr7_+_128784712 | 0.29 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr4_+_144354644 | 0.26 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr15_-_68497657 | 0.26 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr12_-_123201337 | 0.25 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr18_-_52989525 | 0.24 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr12_-_95510743 | 0.22 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr7_+_35756092 | 0.19 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr4_-_25865159 | 0.19 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr7_+_35756186 | 0.19 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr17_-_39684550 | 0.19 |
ENST00000455635.1 ENST00000361566.3 |
KRT19 |
keratin 19 |
| chrX_+_105192423 | 0.19 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr6_+_15246501 | 0.17 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr17_-_41739283 | 0.16 |
ENST00000393661.2 ENST00000318579.4 |
MEOX1 |
mesenchyme homeobox 1 |
| chr5_-_115872142 | 0.16 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_-_124976185 | 0.16 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr13_-_103719196 | 0.15 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr8_-_21669826 | 0.15 |
ENST00000517328.1 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr13_+_109248500 | 0.15 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr9_-_124976154 | 0.15 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr14_-_57277163 | 0.14 |
ENST00000555006.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr7_-_27205136 | 0.14 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr6_+_12290586 | 0.14 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr4_-_87028478 | 0.14 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr14_-_72458326 | 0.14 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr5_-_76935513 | 0.13 |
ENST00000306422.3 |
OTP |
orthopedia homeobox |
| chr17_-_46657473 | 0.13 |
ENST00000332503.5 |
HOXB4 |
homeobox B4 |
| chr4_+_95972822 | 0.13 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr1_+_46152886 | 0.13 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr1_-_200379129 | 0.13 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr2_-_136678123 | 0.12 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr11_+_63606373 | 0.12 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr1_-_200379180 | 0.12 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr2_+_27440229 | 0.12 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_-_81399411 | 0.12 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr13_+_97928395 | 0.11 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr11_+_34663913 | 0.11 |
ENST00000532302.1 |
EHF |
ets homologous factor |
| chrX_+_65384182 | 0.11 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr2_+_166428839 | 0.11 |
ENST00000342316.4 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chrX_+_65382433 | 0.11 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr19_-_19626838 | 0.10 |
ENST00000360913.3 |
TSSK6 |
testis-specific serine kinase 6 |
| chr8_-_20040601 | 0.10 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr8_-_20040638 | 0.10 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chrX_+_65384052 | 0.10 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr20_+_58713514 | 0.10 |
ENST00000432910.1 |
RP5-1043L13.1 |
RP5-1043L13.1 |
| chr13_-_28545276 | 0.10 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chrX_+_14547632 | 0.10 |
ENST00000218075.4 |
GLRA2 |
glycine receptor, alpha 2 |
| chr2_-_51259229 | 0.10 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chr3_+_15045419 | 0.09 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr20_+_42984330 | 0.09 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr2_-_51259292 | 0.09 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chr8_-_109799793 | 0.09 |
ENST00000297459.3 |
TMEM74 |
transmembrane protein 74 |
| chr1_+_209859510 | 0.09 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr18_+_61223393 | 0.09 |
ENST00000269491.1 ENST00000382768.1 |
SERPINB12 |
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr19_+_1440838 | 0.09 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr17_-_38821373 | 0.09 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr14_+_97925151 | 0.09 |
ENST00000554862.1 ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1 |
CTD-2506J14.1 |
| chr3_-_45838011 | 0.09 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr14_-_57272366 | 0.09 |
ENST00000554788.1 ENST00000554845.1 ENST00000408990.3 |
OTX2 |
orthodenticle homeobox 2 |
| chr22_+_39916558 | 0.09 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr9_+_130026756 | 0.08 |
ENST00000314904.5 ENST00000373387.4 |
GARNL3 |
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_79816965 | 0.08 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr12_+_70219052 | 0.08 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr1_+_26485511 | 0.08 |
ENST00000374268.3 |
FAM110D |
family with sequence similarity 110, member D |
| chr1_+_209878182 | 0.08 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr12_-_24102576 | 0.08 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr12_-_18243119 | 0.08 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr15_+_69854027 | 0.07 |
ENST00000498938.2 |
RP11-279F6.1 |
RP11-279F6.1 |
| chr8_-_114449112 | 0.07 |
ENST00000455883.2 ENST00000352409.3 ENST00000297405.5 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr12_+_1800179 | 0.07 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr11_+_34664014 | 0.07 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr13_-_46626847 | 0.07 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr9_-_133814455 | 0.07 |
ENST00000448616.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr8_+_26150628 | 0.06 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr11_-_118305921 | 0.06 |
ENST00000532619.1 |
RP11-770J1.4 |
RP11-770J1.4 |
| chr19_+_4402659 | 0.06 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chrX_-_100604184 | 0.06 |
ENST00000372902.3 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr1_+_110993795 | 0.06 |
ENST00000271331.3 |
PROK1 |
prokineticin 1 |
| chr12_-_71148357 | 0.06 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr15_+_36887069 | 0.06 |
ENST00000566807.1 ENST00000567389.1 ENST00000562877.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr1_+_23345943 | 0.06 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr19_+_36036583 | 0.06 |
ENST00000392205.1 |
TMEM147 |
transmembrane protein 147 |
| chr4_+_169013666 | 0.06 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr12_-_71148413 | 0.06 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr1_+_23345930 | 0.06 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr10_+_114709999 | 0.06 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_52203789 | 0.06 |
ENST00000599343.1 |
AC068987.1 |
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr17_+_7211656 | 0.06 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr3_+_173116225 | 0.06 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr7_-_11871815 | 0.06 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chrM_+_4431 | 0.06 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr14_+_56127989 | 0.06 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr2_-_207023918 | 0.05 |
ENST00000455934.2 ENST00000449699.1 ENST00000454195.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_-_94147385 | 0.05 |
ENST00000260502.6 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr17_+_7461580 | 0.05 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_166930131 | 0.05 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr7_-_108209897 | 0.05 |
ENST00000313516.5 |
THAP5 |
THAP domain containing 5 |
| chr10_-_96829246 | 0.05 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr11_-_132813566 | 0.05 |
ENST00000331898.7 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
| chr15_+_76352178 | 0.05 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr14_-_57277178 | 0.04 |
ENST00000339475.5 ENST00000554559.1 ENST00000555804.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr3_+_63428982 | 0.04 |
ENST00000479198.1 ENST00000460711.1 ENST00000465156.1 |
SYNPR |
synaptoporin |
| chr7_-_81399329 | 0.04 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr19_+_36036477 | 0.04 |
ENST00000222284.5 ENST00000392204.2 |
TMEM147 |
transmembrane protein 147 |
| chr7_-_20826504 | 0.04 |
ENST00000418710.2 ENST00000361443.4 |
SP8 |
Sp8 transcription factor |
| chr7_-_151217001 | 0.04 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr17_+_58018269 | 0.04 |
ENST00000591035.1 |
RP11-178C3.1 |
Uncharacterized protein |
| chr9_-_133814527 | 0.04 |
ENST00000451466.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr3_-_20053741 | 0.04 |
ENST00000389050.4 |
PP2D1 |
protein phosphatase 2C-like domain containing 1 |
| chr1_+_167298281 | 0.04 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr12_-_52779433 | 0.03 |
ENST00000257951.3 |
KRT84 |
keratin 84 |
| chr17_-_77924627 | 0.03 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr14_+_73706308 | 0.03 |
ENST00000554301.1 ENST00000555445.1 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr6_+_152011628 | 0.03 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr18_+_63417864 | 0.03 |
ENST00000536984.2 |
CDH7 |
cadherin 7, type 2 |
| chr16_-_21663950 | 0.03 |
ENST00000268389.4 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr12_+_64173583 | 0.03 |
ENST00000261234.6 |
TMEM5 |
transmembrane protein 5 |
| chr6_-_31745085 | 0.03 |
ENST00000375686.3 ENST00000447450.1 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr6_+_127898312 | 0.03 |
ENST00000329722.7 |
C6orf58 |
chromosome 6 open reading frame 58 |
| chr10_-_13523073 | 0.03 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr4_-_174451370 | 0.03 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr1_+_151739131 | 0.02 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr6_-_31745037 | 0.02 |
ENST00000375688.4 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr15_+_49715293 | 0.02 |
ENST00000267843.4 ENST00000560270.1 |
FGF7 |
fibroblast growth factor 7 |
| chr2_+_177015122 | 0.02 |
ENST00000468418.3 |
HOXD3 |
homeobox D3 |
| chr18_-_74728998 | 0.02 |
ENST00000359645.3 ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP |
myelin basic protein |
| chr2_+_176987088 | 0.02 |
ENST00000249499.6 |
HOXD9 |
homeobox D9 |
| chr12_-_111358372 | 0.02 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr3_-_57326704 | 0.02 |
ENST00000487349.1 ENST00000389601.3 |
ASB14 |
ankyrin repeat and SOCS box containing 14 |
| chr8_+_76452097 | 0.02 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr9_+_95736758 | 0.02 |
ENST00000337352.6 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
| chr5_-_36301984 | 0.02 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr9_-_16705069 | 0.02 |
ENST00000471301.2 |
BNC2 |
basonuclin 2 |
| chr2_-_209010874 | 0.01 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr16_+_67906919 | 0.01 |
ENST00000358933.5 |
EDC4 |
enhancer of mRNA decapping 4 |
| chr6_-_91296737 | 0.01 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr12_+_130646999 | 0.01 |
ENST00000539839.1 ENST00000229030.4 |
FZD10 |
frizzled family receptor 10 |
| chrX_-_10851762 | 0.01 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr19_-_19626351 | 0.01 |
ENST00000585580.3 |
TSSK6 |
testis-specific serine kinase 6 |
| chr7_-_81399355 | 0.01 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_128293323 | 0.01 |
ENST00000389524.4 ENST00000428314.1 |
MYO7B |
myosin VIIB |
| chr3_-_51813009 | 0.01 |
ENST00000398780.3 |
IQCF6 |
IQ motif containing F6 |
| chr5_-_169626104 | 0.01 |
ENST00000520275.1 ENST00000506431.2 |
CTB-27N1.1 |
CTB-27N1.1 |
| chr1_-_9129631 | 0.01 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr10_-_24770632 | 0.01 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr7_-_93519471 | 0.01 |
ENST00000451238.1 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr8_-_59412717 | 0.01 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr15_-_54025300 | 0.01 |
ENST00000559418.1 |
WDR72 |
WD repeat domain 72 |
| chr6_-_91296602 | 0.01 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr1_+_75600567 | 0.01 |
ENST00000356261.3 |
LHX8 |
LIM homeobox 8 |
| chr7_+_114055052 | 0.00 |
ENST00000462331.1 ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2 |
forkhead box P2 |
| chr1_+_66999268 | 0.00 |
ENST00000371039.1 ENST00000424320.1 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr2_-_157198860 | 0.00 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_137815524 | 0.00 |
ENST00000367734.2 |
OLIG3 |
oligodendrocyte transcription factor 3 |
| chr10_+_26505179 | 0.00 |
ENST00000376261.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr5_+_159656437 | 0.00 |
ENST00000402432.3 |
FABP6 |
fatty acid binding protein 6, ileal |
| chr6_+_26156551 | 0.00 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr1_+_87794150 | 0.00 |
ENST00000370544.5 |
LMO4 |
LIM domain only 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.3 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |