Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for HOXD4
Z-value: 0.48
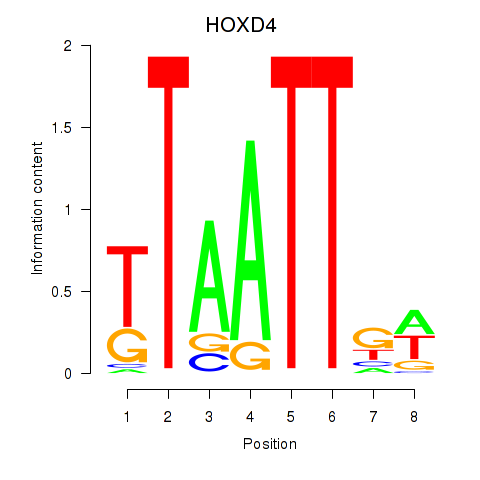
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | HOXD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | 0.18 | 6.8e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 1.00 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr1_+_209602771 | 0.92 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr6_+_130339710 | 0.48 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr2_-_165424973 | 0.45 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr7_-_22233442 | 0.39 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_-_136847099 | 0.35 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr11_+_128563652 | 0.28 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_69313145 | 0.27 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr12_+_41831485 | 0.26 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr3_+_142342228 | 0.25 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr5_+_66300446 | 0.25 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_31361543 | 0.23 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_186798073 | 0.22 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr4_-_143227088 | 0.22 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_16793160 | 0.22 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr4_-_153601136 | 0.22 |
ENST00000504064.1 ENST00000304385.3 |
TMEM154 |
transmembrane protein 154 |
| chr4_-_143226979 | 0.21 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr3_+_111718173 | 0.21 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr12_-_39734783 | 0.21 |
ENST00000552961.1 |
KIF21A |
kinesin family member 21A |
| chr14_+_29236269 | 0.20 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr4_-_130014609 | 0.19 |
ENST00000511426.1 |
SCLT1 |
sodium channel and clathrin linker 1 |
| chr4_-_130014705 | 0.19 |
ENST00000503401.1 |
SCLT1 |
sodium channel and clathrin linker 1 |
| chr17_-_26733179 | 0.16 |
ENST00000440501.1 ENST00000321666.5 |
SLC46A1 |
solute carrier family 46 (folate transporter), member 1 |
| chr11_-_72492878 | 0.15 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr21_+_43823983 | 0.15 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr6_+_53659746 | 0.15 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr18_-_74839891 | 0.15 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr7_-_25268104 | 0.15 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr2_+_191045656 | 0.14 |
ENST00000443551.2 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chr3_-_190167571 | 0.13 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr1_-_120935894 | 0.13 |
ENST00000369383.4 ENST00000369384.4 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr17_-_64225508 | 0.13 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_204801471 | 0.12 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr4_-_76944621 | 0.12 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr8_-_57026541 | 0.12 |
ENST00000311923.1 |
MOS |
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr17_-_10452929 | 0.12 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr8_-_124279627 | 0.12 |
ENST00000357082.4 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
| chr4_+_155484155 | 0.11 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr13_+_73629107 | 0.11 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr7_-_137028534 | 0.11 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr21_-_40984704 | 0.11 |
ENST00000329618.6 |
C21orf88 |
chromosome 21 open reading frame 88 |
| chr7_-_137028498 | 0.10 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr2_+_32502952 | 0.10 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr4_-_130014532 | 0.10 |
ENST00000506368.1 ENST00000439369.2 ENST00000503215.1 |
SCLT1 |
sodium channel and clathrin linker 1 |
| chr8_+_50822344 | 0.10 |
ENST00000518864.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr2_-_99871570 | 0.10 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr2_+_234826016 | 0.10 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_-_78809950 | 0.10 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr3_-_126327398 | 0.10 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr9_+_470288 | 0.09 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr12_-_91451758 | 0.09 |
ENST00000266719.3 |
KERA |
keratocan |
| chr9_+_130026756 | 0.09 |
ENST00000314904.5 ENST00000373387.4 |
GARNL3 |
GTPase activating Rap/RanGAP domain-like 3 |
| chr12_-_81763184 | 0.09 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_-_191000172 | 0.09 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr15_-_54025300 | 0.09 |
ENST00000559418.1 |
WDR72 |
WD repeat domain 72 |
| chr18_-_19994830 | 0.09 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr13_-_45768841 | 0.09 |
ENST00000379108.1 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
| chr18_-_19283649 | 0.09 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr6_-_53013620 | 0.09 |
ENST00000259803.7 |
GCM1 |
glial cells missing homolog 1 (Drosophila) |
| chr20_+_42136308 | 0.08 |
ENST00000434666.1 ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
| chr22_+_23248512 | 0.08 |
ENST00000390325.2 |
IGLC3 |
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr14_-_72458326 | 0.08 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr6_-_44281043 | 0.08 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr5_-_13944652 | 0.08 |
ENST00000265104.4 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
| chr2_-_160473114 | 0.08 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr3_+_15045419 | 0.08 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr7_-_121944491 | 0.08 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr4_-_130014729 | 0.08 |
ENST00000281142.5 ENST00000434680.1 |
SCLT1 |
sodium channel and clathrin linker 1 |
| chr3_-_137851220 | 0.08 |
ENST00000236709.3 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
| chr3_+_97483572 | 0.07 |
ENST00000335979.2 ENST00000394206.1 |
ARL6 |
ADP-ribosylation factor-like 6 |
| chr22_-_29107919 | 0.07 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr2_+_90198535 | 0.07 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr16_+_84801852 | 0.07 |
ENST00000569925.1 ENST00000567526.1 |
USP10 |
ubiquitin specific peptidase 10 |
| chr1_-_36948879 | 0.07 |
ENST00000373106.1 ENST00000373104.1 ENST00000373103.1 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr12_+_21168630 | 0.07 |
ENST00000421593.2 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_+_42846443 | 0.07 |
ENST00000410070.2 ENST00000431473.3 |
RIMKLA |
ribosomal modification protein rimK-like family member A |
| chr1_-_87380002 | 0.07 |
ENST00000331835.5 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr2_-_160472952 | 0.07 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr1_-_28384598 | 0.07 |
ENST00000373864.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr1_+_182419261 | 0.06 |
ENST00000294854.8 ENST00000542961.1 |
RGSL1 |
regulator of G-protein signaling like 1 |
| chr20_+_30063067 | 0.06 |
ENST00000201979.2 |
REM1 |
RAS (RAD and GEM)-like GTP-binding 1 |
| chr20_-_13971255 | 0.06 |
ENST00000284951.5 ENST00000378072.5 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr3_+_140981456 | 0.06 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr16_+_24549014 | 0.06 |
ENST00000564314.1 ENST00000567686.1 |
RBBP6 |
retinoblastoma binding protein 6 |
| chr15_-_88799661 | 0.06 |
ENST00000360948.2 ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr3_+_63638339 | 0.06 |
ENST00000343837.3 ENST00000469440.1 |
SNTN |
sentan, cilia apical structure protein |
| chr1_-_92952433 | 0.06 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr7_-_104909435 | 0.06 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr2_-_225434538 | 0.06 |
ENST00000409096.1 |
CUL3 |
cullin 3 |
| chr1_+_197237352 | 0.06 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr12_-_86650045 | 0.06 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_196313239 | 0.06 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr1_+_115572415 | 0.06 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr18_-_61311485 | 0.05 |
ENST00000436264.1 ENST00000356424.6 ENST00000341074.5 |
SERPINB4 |
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr20_-_29978383 | 0.05 |
ENST00000339144.3 ENST00000376321.3 |
DEFB119 |
defensin, beta 119 |
| chr9_-_128246769 | 0.05 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr2_-_27579842 | 0.05 |
ENST00000423998.1 ENST00000264720.3 |
GTF3C2 |
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr10_-_50970322 | 0.05 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr13_+_33160553 | 0.05 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr4_+_146560245 | 0.05 |
ENST00000541599.1 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr12_+_112279782 | 0.05 |
ENST00000550735.2 |
MAPKAPK5 |
mitogen-activated protein kinase-activated protein kinase 5 |
| chr1_+_67632083 | 0.05 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chrX_+_130192318 | 0.05 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr6_+_123317116 | 0.05 |
ENST00000275162.5 |
CLVS2 |
clavesin 2 |
| chr15_+_80364901 | 0.05 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr4_-_39979576 | 0.05 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chrX_-_10645773 | 0.05 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr4_-_103940791 | 0.05 |
ENST00000510559.1 ENST00000394789.3 ENST00000296422.7 |
SLC9B1 |
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr1_+_109256067 | 0.05 |
ENST00000271311.2 |
FNDC7 |
fibronectin type III domain containing 7 |
| chr12_-_118628350 | 0.05 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr7_+_112063192 | 0.05 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr5_-_39274617 | 0.05 |
ENST00000510188.1 |
FYB |
FYN binding protein |
| chr2_-_159237472 | 0.05 |
ENST00000409187.1 |
CCDC148 |
coiled-coil domain containing 148 |
| chr16_+_87636474 | 0.04 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chr17_-_41132410 | 0.04 |
ENST00000409446.3 ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr1_-_87379785 | 0.04 |
ENST00000401030.3 ENST00000370554.1 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr20_+_18447771 | 0.04 |
ENST00000377603.4 |
POLR3F |
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr4_+_70796784 | 0.04 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr3_-_113897545 | 0.04 |
ENST00000467632.1 |
DRD3 |
dopamine receptor D3 |
| chrX_-_110655306 | 0.04 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr4_+_155484103 | 0.04 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr1_-_190446759 | 0.04 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr17_-_29624343 | 0.04 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr1_-_231560790 | 0.04 |
ENST00000366641.3 |
EGLN1 |
egl-9 family hypoxia-inducible factor 1 |
| chr13_-_24007815 | 0.04 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr4_-_26492076 | 0.04 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr11_-_89224638 | 0.04 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_64684672 | 0.03 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr3_-_4508925 | 0.03 |
ENST00000534863.1 ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1 |
sulfatase modifying factor 1 |
| chr2_+_27886330 | 0.03 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr6_+_142468361 | 0.03 |
ENST00000367630.4 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
| chr5_-_111754948 | 0.03 |
ENST00000261486.5 |
EPB41L4A |
erythrocyte membrane protein band 4.1 like 4A |
| chr21_-_27423339 | 0.03 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr15_+_36887069 | 0.03 |
ENST00000566807.1 ENST00000567389.1 ENST00000562877.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr17_+_41132564 | 0.03 |
ENST00000361677.1 ENST00000589705.1 |
RUNDC1 |
RUN domain containing 1 |
| chr2_+_234590556 | 0.03 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr3_+_51575596 | 0.03 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr10_-_52008313 | 0.03 |
ENST00000329428.6 ENST00000395526.4 ENST00000447815.1 |
ASAH2 |
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr6_-_32157947 | 0.03 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr1_+_87380299 | 0.03 |
ENST00000370551.4 ENST00000370550.5 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
| chr4_+_88532028 | 0.03 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr2_+_176981307 | 0.03 |
ENST00000249501.4 |
HOXD10 |
homeobox D10 |
| chr19_+_30433110 | 0.03 |
ENST00000542441.2 ENST00000392271.1 |
URI1 |
URI1, prefoldin-like chaperone |
| chr3_+_173116225 | 0.03 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr8_+_24151553 | 0.02 |
ENST00000265769.4 ENST00000540823.1 ENST00000397649.3 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr15_-_64386120 | 0.02 |
ENST00000300030.3 |
FAM96A |
family with sequence similarity 96, member A |
| chr12_-_49245936 | 0.02 |
ENST00000308025.3 |
DDX23 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr2_-_163695238 | 0.02 |
ENST00000328032.4 |
KCNH7 |
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr18_-_51750948 | 0.02 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chrX_+_139791917 | 0.02 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr17_+_9479944 | 0.02 |
ENST00000396219.3 ENST00000352665.5 |
WDR16 |
WD repeat domain 16 |
| chr17_+_3118915 | 0.02 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chr9_+_273038 | 0.02 |
ENST00000487230.1 ENST00000469391.1 |
DOCK8 |
dedicator of cytokinesis 8 |
| chr12_-_112279694 | 0.02 |
ENST00000443596.1 ENST00000442119.1 |
MAPKAPK5-AS1 |
MAPKAPK5 antisense RNA 1 |
| chr6_-_26108355 | 0.02 |
ENST00000338379.4 |
HIST1H1T |
histone cluster 1, H1t |
| chr14_-_99947168 | 0.02 |
ENST00000331768.5 |
SETD3 |
SET domain containing 3 |
| chr7_+_135611542 | 0.02 |
ENST00000416501.1 |
AC015987.2 |
AC015987.2 |
| chr6_-_26250835 | 0.02 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr15_-_64385981 | 0.02 |
ENST00000557835.1 ENST00000380290.3 ENST00000559950.1 |
FAM96A |
family with sequence similarity 96, member A |
| chr7_+_77428149 | 0.02 |
ENST00000415251.2 ENST00000275575.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr4_-_89951028 | 0.02 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr5_+_69321074 | 0.02 |
ENST00000380751.5 ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B |
small EDRK-rich factor 1B (centromeric) |
| chr20_-_43729750 | 0.02 |
ENST00000537075.1 ENST00000306117.1 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr8_-_27941380 | 0.02 |
ENST00000413272.2 ENST00000341513.6 |
NUGGC |
nuclear GTPase, germinal center associated |
| chr20_+_32150140 | 0.02 |
ENST00000344201.3 ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr11_-_89224139 | 0.01 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr8_+_24151620 | 0.01 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr5_-_55412774 | 0.01 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr6_-_49712147 | 0.01 |
ENST00000433368.2 ENST00000354620.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr2_+_187371440 | 0.01 |
ENST00000445547.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr2_+_224822121 | 0.01 |
ENST00000258383.3 |
MRPL44 |
mitochondrial ribosomal protein L44 |
| chr4_-_70505358 | 0.01 |
ENST00000457664.2 ENST00000604629.1 ENST00000604021.1 |
UGT2A2 |
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr17_-_3030875 | 0.01 |
ENST00000328890.2 |
OR1G1 |
olfactory receptor, family 1, subfamily G, member 1 |
| chr3_-_4927447 | 0.01 |
ENST00000449914.1 |
AC018816.3 |
Uncharacterized protein |
| chr11_-_40315640 | 0.01 |
ENST00000278198.2 |
LRRC4C |
leucine rich repeat containing 4C |
| chr11_-_89223883 | 0.01 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_-_11623804 | 0.01 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr11_-_121986923 | 0.01 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr7_-_77427676 | 0.01 |
ENST00000257663.3 |
TMEM60 |
transmembrane protein 60 |
| chr15_-_56209306 | 0.01 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chrX_+_56590002 | 0.01 |
ENST00000338222.5 |
UBQLN2 |
ubiquilin 2 |
| chr4_-_69111401 | 0.01 |
ENST00000332644.5 |
TMPRSS11B |
transmembrane protease, serine 11B |
| chr14_+_78174414 | 0.01 |
ENST00000557342.1 ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP |
SRA stem-loop interacting RNA binding protein |
| chr17_-_60142609 | 0.01 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr16_-_21663950 | 0.01 |
ENST00000268389.4 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr5_+_59783941 | 0.00 |
ENST00000506884.1 ENST00000504876.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr14_+_99947715 | 0.00 |
ENST00000389879.5 ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK |
cyclin K |
| chr10_+_18629628 | 0.00 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_+_101460882 | 0.00 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr2_+_207024306 | 0.00 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_117963209 | 0.00 |
ENST00000449370.2 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0034392 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |