Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for HSF1
Z-value: 2.04
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | HSF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | 0.25 | 5.5e-01 | Click! |
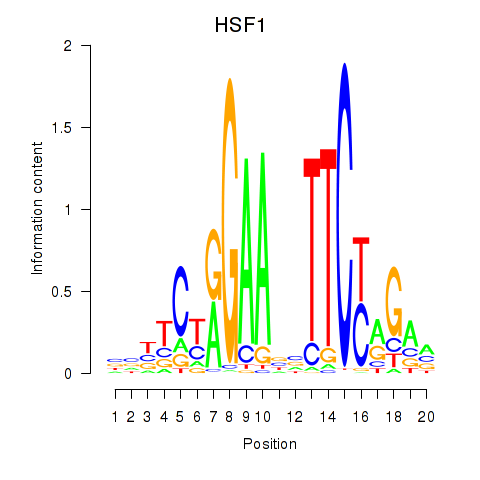
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51504411 | 2.12 |
ENST00000593490.1 |
KLK8 |
kallikrein-related peptidase 8 |
| chr19_-_51522955 | 2.08 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr19_-_36004543 | 1.98 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr2_+_102624977 | 1.80 |
ENST00000441002.1 |
IL1R2 |
interleukin 1 receptor, type II |
| chr1_+_35247859 | 1.73 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr19_-_6720686 | 1.69 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr2_+_47596287 | 1.64 |
ENST00000263735.4 |
EPCAM |
epithelial cell adhesion molecule |
| chr18_+_21452964 | 1.62 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452804 | 1.56 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr6_+_47666275 | 1.48 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr19_+_751122 | 1.45 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr17_-_39677971 | 1.38 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr6_-_136788001 | 1.33 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr19_-_51456198 | 1.32 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr1_-_153363452 | 1.32 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr19_-_51523412 | 1.30 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr19_-_51523275 | 1.21 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr8_+_124194752 | 1.20 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr19_-_51456344 | 1.18 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr20_+_43803517 | 1.17 |
ENST00000243924.3 |
PI3 |
peptidase inhibitor 3, skin-derived |
| chr1_+_28261621 | 1.11 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_10471836 | 1.11 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr1_+_28261492 | 1.09 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr12_-_52911718 | 1.09 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr1_+_152957707 | 1.08 |
ENST00000368762.1 |
SPRR1A |
small proline-rich protein 1A |
| chr1_+_15256230 | 1.03 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr10_-_116444371 | 1.01 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr2_-_238499303 | 1.00 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr1_+_43996518 | 0.99 |
ENST00000359947.4 ENST00000438120.1 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
| chr19_-_49567124 | 0.93 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr2_-_238499131 | 0.90 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr15_-_72490114 | 0.89 |
ENST00000309731.7 |
GRAMD2 |
GRAM domain containing 2 |
| chr17_-_3595181 | 0.88 |
ENST00000552050.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr1_+_220960033 | 0.82 |
ENST00000366910.5 |
MARC1 |
mitochondrial amidoxime reducing component 1 |
| chr6_-_170599561 | 0.76 |
ENST00000366756.3 |
DLL1 |
delta-like 1 (Drosophila) |
| chr17_-_39928106 | 0.75 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr2_+_85360499 | 0.72 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr7_-_139763521 | 0.70 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr1_-_24469602 | 0.69 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr20_+_61287711 | 0.68 |
ENST00000370507.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chr17_-_76123101 | 0.67 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
| chr17_-_39661849 | 0.65 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr1_-_147232669 | 0.63 |
ENST00000369237.1 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr11_-_88070896 | 0.63 |
ENST00000529974.1 ENST00000527018.1 |
CTSC |
cathepsin C |
| chr19_-_55660561 | 0.62 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr20_+_1875378 | 0.57 |
ENST00000356025.3 |
SIRPA |
signal-regulatory protein alpha |
| chr20_+_1875110 | 0.57 |
ENST00000400068.3 |
SIRPA |
signal-regulatory protein alpha |
| chr16_-_85784557 | 0.57 |
ENST00000602675.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr3_+_124223586 | 0.56 |
ENST00000393496.1 |
KALRN |
kalirin, RhoGEF kinase |
| chr19_+_49055332 | 0.56 |
ENST00000201586.2 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
| chrX_+_147062844 | 0.56 |
ENST00000370467.3 |
FMR1NB |
fragile X mental retardation 1 neighbor |
| chr4_+_76481258 | 0.55 |
ENST00000311623.4 ENST00000435974.2 |
C4orf26 |
chromosome 4 open reading frame 26 |
| chr1_+_28261533 | 0.54 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr20_-_18038521 | 0.53 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr6_+_35265586 | 0.52 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr6_-_32784687 | 0.51 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr6_-_33041378 | 0.50 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr3_+_130612803 | 0.50 |
ENST00000510168.1 ENST00000508532.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr11_+_59480899 | 0.49 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr8_-_81083890 | 0.49 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chr12_+_115800817 | 0.47 |
ENST00000547948.1 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
| chr8_+_8559406 | 0.47 |
ENST00000519106.1 |
CLDN23 |
claudin 23 |
| chr1_+_156117149 | 0.46 |
ENST00000435124.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr9_+_470288 | 0.45 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr20_+_10015678 | 0.45 |
ENST00000378392.1 ENST00000378380.3 |
ANKEF1 |
ankyrin repeat and EF-hand domain containing 1 |
| chr11_+_33563821 | 0.43 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chrX_-_100662881 | 0.43 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr7_+_130020932 | 0.43 |
ENST00000484324.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr15_+_75491213 | 0.42 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr3_+_130613001 | 0.42 |
ENST00000504948.1 ENST00000513801.1 ENST00000505072.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_96722539 | 0.42 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr8_-_81083341 | 0.42 |
ENST00000519303.2 |
TPD52 |
tumor protein D52 |
| chr19_+_1065922 | 0.41 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chrX_-_15683147 | 0.41 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr5_-_172036436 | 0.40 |
ENST00000601856.1 |
AC027309.1 |
AC027309.1 |
| chr8_-_81083731 | 0.40 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chrX_-_152989531 | 0.40 |
ENST00000458587.2 ENST00000416815.1 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr12_+_2904102 | 0.40 |
ENST00000001008.4 |
FKBP4 |
FK506 binding protein 4, 59kDa |
| chr19_-_55672037 | 0.39 |
ENST00000588076.1 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chr2_-_96701722 | 0.39 |
ENST00000434632.1 |
GPAT2 |
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr5_-_175964366 | 0.39 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr4_-_48908737 | 0.38 |
ENST00000381464.2 |
OCIAD2 |
OCIA domain containing 2 |
| chr19_+_8117881 | 0.38 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr4_-_25865159 | 0.38 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr2_+_113735575 | 0.38 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr8_-_56685859 | 0.38 |
ENST00000523423.1 ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68 |
transmembrane protein 68 |
| chrX_-_152989798 | 0.38 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr1_+_212782012 | 0.38 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr2_+_231577532 | 0.36 |
ENST00000258418.5 |
CAB39 |
calcium binding protein 39 |
| chr1_-_10856694 | 0.36 |
ENST00000377022.3 ENST00000344008.5 |
CASZ1 |
castor zinc finger 1 |
| chrX_+_30233668 | 0.36 |
ENST00000378988.4 |
MAGEB2 |
melanoma antigen family B, 2 |
| chr11_-_89956227 | 0.36 |
ENST00000457199.2 ENST00000530765.1 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr19_-_17445613 | 0.36 |
ENST00000159087.4 |
ANO8 |
anoctamin 8 |
| chr4_-_48908822 | 0.36 |
ENST00000508632.1 |
OCIAD2 |
OCIA domain containing 2 |
| chr4_-_48908805 | 0.35 |
ENST00000273860.4 |
OCIAD2 |
OCIA domain containing 2 |
| chr7_+_141478242 | 0.35 |
ENST00000247881.2 |
TAS2R4 |
taste receptor, type 2, member 4 |
| chr1_+_196857144 | 0.35 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr11_+_112046190 | 0.35 |
ENST00000357685.5 ENST00000393032.2 ENST00000361053.4 |
BCO2 |
beta-carotene oxygenase 2 |
| chr5_+_141016969 | 0.34 |
ENST00000518856.1 |
RELL2 |
RELT-like 2 |
| chr18_+_60382672 | 0.34 |
ENST00000400316.4 ENST00000262719.5 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
| chr1_+_67773044 | 0.34 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr20_+_18794370 | 0.34 |
ENST00000377428.2 |
SCP2D1 |
SCP2 sterol-binding domain containing 1 |
| chr1_-_247921982 | 0.34 |
ENST00000408896.2 |
OR1C1 |
olfactory receptor, family 1, subfamily C, member 1 |
| chr3_-_123411191 | 0.34 |
ENST00000354792.5 ENST00000508240.1 |
MYLK |
myosin light chain kinase |
| chr19_-_14629224 | 0.34 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr17_+_53828333 | 0.33 |
ENST00000268896.5 |
PCTP |
phosphatidylcholine transfer protein |
| chr3_+_32433154 | 0.33 |
ENST00000334983.5 ENST00000349718.4 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr3_+_32433363 | 0.32 |
ENST00000465248.1 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr3_+_130613226 | 0.32 |
ENST00000509662.1 ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_141016508 | 0.32 |
ENST00000444782.1 ENST00000521367.1 ENST00000297164.3 |
RELL2 |
RELT-like 2 |
| chr12_+_50366620 | 0.32 |
ENST00000315520.5 |
AQP6 |
aquaporin 6, kidney specific |
| chr19_-_2051223 | 0.32 |
ENST00000309340.7 ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chrX_+_49020882 | 0.31 |
ENST00000454342.1 |
MAGIX |
MAGI family member, X-linked |
| chr14_+_22977587 | 0.31 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr2_+_109403193 | 0.31 |
ENST00000412964.2 ENST00000295124.4 |
CCDC138 |
coiled-coil domain containing 138 |
| chr7_+_43622664 | 0.31 |
ENST00000319357.5 |
STK17A |
serine/threonine kinase 17a |
| chr19_+_8117636 | 0.31 |
ENST00000253451.4 ENST00000315626.4 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr9_+_139847347 | 0.31 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr17_-_54911250 | 0.30 |
ENST00000575658.1 ENST00000397861.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr15_+_68871308 | 0.30 |
ENST00000261861.5 |
CORO2B |
coronin, actin binding protein, 2B |
| chr21_+_39644172 | 0.30 |
ENST00000398932.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_206138457 | 0.30 |
ENST00000367128.3 ENST00000431655.2 |
FAM72A |
family with sequence similarity 72, member A |
| chr14_-_77787198 | 0.30 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr17_+_54911444 | 0.29 |
ENST00000284061.3 ENST00000572810.1 |
DGKE |
diacylglycerol kinase, epsilon 64kDa |
| chr22_+_38453207 | 0.29 |
ENST00000404072.3 ENST00000424694.1 |
PICK1 |
protein interacting with PRKCA 1 |
| chr21_+_39644214 | 0.29 |
ENST00000438657.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_173472607 | 0.28 |
ENST00000303177.3 ENST00000519867.1 |
NSG2 |
Neuron-specific protein family member 2 |
| chr12_+_51632666 | 0.28 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr17_+_53828381 | 0.28 |
ENST00000576183.1 |
PCTP |
phosphatidylcholine transfer protein |
| chr4_+_41937131 | 0.28 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr6_-_105627735 | 0.28 |
ENST00000254765.3 |
POPDC3 |
popeye domain containing 3 |
| chr18_-_19284724 | 0.28 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr16_+_85942594 | 0.28 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr11_-_89956461 | 0.28 |
ENST00000320585.6 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr2_-_9143786 | 0.27 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr17_-_62208169 | 0.27 |
ENST00000606895.1 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
| chr11_-_47869865 | 0.27 |
ENST00000530326.1 ENST00000532747.1 |
NUP160 |
nucleoporin 160kDa |
| chr6_+_31795506 | 0.27 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr1_+_110577229 | 0.27 |
ENST00000369795.3 ENST00000369794.2 |
STRIP1 |
striatin interacting protein 1 |
| chr1_+_196743912 | 0.27 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr1_+_212738676 | 0.27 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr11_+_93861993 | 0.27 |
ENST00000227638.3 ENST00000436171.2 |
PANX1 |
pannexin 1 |
| chr1_+_120839005 | 0.27 |
ENST00000369390.3 ENST00000452190.1 |
FAM72B |
family with sequence similarity 72, member B |
| chr11_-_104840093 | 0.27 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr22_+_38453378 | 0.26 |
ENST00000437453.1 ENST00000356976.3 |
PICK1 |
protein interacting with PRKCA 1 |
| chr7_-_3083573 | 0.26 |
ENST00000396946.4 |
CARD11 |
caspase recruitment domain family, member 11 |
| chr9_-_34372830 | 0.26 |
ENST00000379142.3 |
KIAA1161 |
KIAA1161 |
| chr22_+_48972118 | 0.26 |
ENST00000358295.5 |
FAM19A5 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr11_+_69061594 | 0.26 |
ENST00000441339.2 ENST00000308946.3 ENST00000535407.1 |
MYEOV |
myeloma overexpressed |
| chr3_+_130569429 | 0.26 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_-_42526802 | 0.25 |
ENST00000359033.4 ENST00000389970.3 ENST00000360608.5 |
CYP2D6 |
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr14_+_39734482 | 0.25 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr10_-_125851961 | 0.25 |
ENST00000346248.5 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr17_+_7258442 | 0.25 |
ENST00000389982.4 ENST00000576060.1 ENST00000330767.4 |
TMEM95 |
transmembrane protein 95 |
| chr2_+_11674213 | 0.25 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chrX_-_154563889 | 0.25 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr11_-_47870019 | 0.25 |
ENST00000378460.2 |
NUP160 |
nucleoporin 160kDa |
| chr19_+_58038683 | 0.25 |
ENST00000240719.3 ENST00000376233.3 ENST00000594943.1 ENST00000602149.1 |
ZNF549 |
zinc finger protein 549 |
| chr12_+_66696322 | 0.25 |
ENST00000247815.4 |
HELB |
helicase (DNA) B |
| chr16_-_57831914 | 0.25 |
ENST00000421376.2 |
KIFC3 |
kinesin family member C3 |
| chr1_+_196743943 | 0.24 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr6_+_73331520 | 0.24 |
ENST00000342056.2 ENST00000355194.4 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr21_-_43346790 | 0.24 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr19_-_54876558 | 0.24 |
ENST00000391742.2 ENST00000434277.2 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
| chr17_+_56315936 | 0.24 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr19_+_39421556 | 0.24 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr3_+_130569592 | 0.24 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_-_167369612 | 0.23 |
ENST00000507747.1 |
RP11-514O12.4 |
RP11-514O12.4 |
| chr21_+_42733870 | 0.23 |
ENST00000330714.3 ENST00000436410.1 ENST00000435611.1 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
| chr19_-_16653226 | 0.23 |
ENST00000198939.6 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr3_-_59035673 | 0.23 |
ENST00000491845.1 ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67 |
chromosome 3 open reading frame 67 |
| chr2_+_173420697 | 0.23 |
ENST00000282077.3 ENST00000392571.2 ENST00000410055.1 |
PDK1 |
pyruvate dehydrogenase kinase, isozyme 1 |
| chr7_-_150780487 | 0.23 |
ENST00000482202.1 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr17_+_40714092 | 0.23 |
ENST00000420359.1 ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY |
CoA synthase |
| chr6_+_4890226 | 0.22 |
ENST00000343762.5 |
CDYL |
chromodomain protein, Y-like |
| chr20_+_55904815 | 0.22 |
ENST00000371263.3 ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11 |
SPO11 meiotic protein covalently bound to DSB |
| chr7_-_111032971 | 0.22 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr19_-_14117074 | 0.22 |
ENST00000588885.1 ENST00000254325.4 |
RFX1 |
regulatory factor X, 1 (influences HLA class II expression) |
| chr3_-_183735731 | 0.22 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_-_12184159 | 0.22 |
ENST00000312019.2 |
RP11-276H1.3 |
RP11-276H1.3 |
| chr19_-_16653325 | 0.22 |
ENST00000546361.2 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr1_-_143913143 | 0.22 |
ENST00000400889.1 |
FAM72D |
family with sequence similarity 72, member D |
| chrX_-_72434628 | 0.21 |
ENST00000536638.1 ENST00000373517.3 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
| chr6_+_31465849 | 0.21 |
ENST00000399150.3 |
MICB |
MHC class I polypeptide-related sequence B |
| chr19_-_10628117 | 0.21 |
ENST00000333430.4 |
S1PR5 |
sphingosine-1-phosphate receptor 5 |
| chr6_-_46620522 | 0.21 |
ENST00000275016.2 |
CYP39A1 |
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr9_-_86955598 | 0.21 |
ENST00000376238.4 |
SLC28A3 |
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr1_+_111415757 | 0.21 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr17_-_17485731 | 0.21 |
ENST00000395783.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr19_+_45349630 | 0.21 |
ENST00000252483.5 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr15_-_83378611 | 0.21 |
ENST00000542200.1 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
| chr9_+_74764278 | 0.21 |
ENST00000238018.4 ENST00000376989.3 |
GDA |
guanine deaminase |
| chr3_+_44916098 | 0.21 |
ENST00000296125.4 |
TGM4 |
transglutaminase 4 |
| chr12_+_70132632 | 0.21 |
ENST00000378815.6 ENST00000483530.2 ENST00000325555.9 |
RAB3IP |
RAB3A interacting protein |
| chr15_+_34394257 | 0.21 |
ENST00000397766.2 |
PGBD4 |
piggyBac transposable element derived 4 |
| chr17_-_56350797 | 0.21 |
ENST00000577220.1 |
MPO |
myeloperoxidase |
| chr12_+_133287392 | 0.21 |
ENST00000317555.2 ENST00000498926.2 |
PGAM5 |
phosphoglycerate mutase family member 5 |
| chr1_+_166958497 | 0.20 |
ENST00000367870.2 |
MAEL |
maelstrom spermatogenic transposon silencer |
| chr11_+_34645791 | 0.20 |
ENST00000529527.1 ENST00000531728.1 ENST00000525253.1 |
EHF |
ets homologous factor |
| chr22_-_26961328 | 0.20 |
ENST00000398110.2 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 1.8 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.4 | 2.5 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 1.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 2.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 2.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 0.3 | GO:0090346 | cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.3 | 0.8 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.9 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 0.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.2 | 0.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 2.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.6 | GO:0010652 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.2 | 1.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 1.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 0.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.2 | GO:0090288 | negative regulation of cellular response to growth factor stimulus(GO:0090288) |
| 0.2 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 4.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.8 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 1.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.5 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.6 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.2 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.2 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 0.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.3 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.5 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 1.2 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.2 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.6 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.3 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 3.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 2.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.8 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 3.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 2.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 1.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 2.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 2.1 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 2.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.8 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 0.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 2.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.3 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.2 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 3.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III transcription factor binding(GO:0001025) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 9.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0019239 | deaminase activity(GO:0019239) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 4.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |