Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
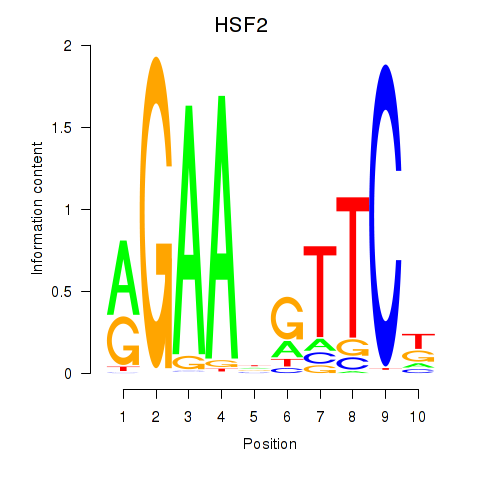
Results for HSF2
Z-value: 0.55
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | HSF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg19_v2_chr6_+_122720681_122720713 | -0.28 | 5.0e-01 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_20805109 | 0.48 |
ENST00000241124.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr4_+_77356248 | 0.34 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr17_-_56494882 | 0.31 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494713 | 0.31 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494908 | 0.31 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr18_-_47376197 | 0.28 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr18_-_52989217 | 0.24 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr19_-_49567124 | 0.22 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr18_-_52989525 | 0.21 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr4_-_22517620 | 0.18 |
ENST00000502482.1 ENST00000334304.5 |
GPR125 |
G protein-coupled receptor 125 |
| chr2_+_162272605 | 0.17 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr14_-_23624511 | 0.16 |
ENST00000529705.2 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr6_+_106534192 | 0.16 |
ENST00000369091.2 ENST00000369096.4 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr17_-_39661849 | 0.16 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr11_+_1856034 | 0.15 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr8_-_81083341 | 0.15 |
ENST00000519303.2 |
TPD52 |
tumor protein D52 |
| chr6_+_29364416 | 0.15 |
ENST00000383555.2 |
OR12D2 |
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr6_-_53013620 | 0.14 |
ENST00000259803.7 |
GCM1 |
glial cells missing homolog 1 (Drosophila) |
| chr11_+_1855645 | 0.14 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr4_-_153601136 | 0.14 |
ENST00000504064.1 ENST00000304385.3 |
TMEM154 |
transmembrane protein 154 |
| chr3_-_129407535 | 0.14 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr8_+_134125727 | 0.14 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr16_-_30125177 | 0.14 |
ENST00000406256.3 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chrX_-_51239425 | 0.14 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chrX_-_152989531 | 0.14 |
ENST00000458587.2 ENST00000416815.1 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr12_-_14721283 | 0.14 |
ENST00000240617.5 |
PLBD1 |
phospholipase B domain containing 1 |
| chr18_-_19284724 | 0.13 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr19_-_14629224 | 0.12 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_-_89956227 | 0.12 |
ENST00000457199.2 ENST00000530765.1 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr7_+_130126012 | 0.11 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr19_+_14142535 | 0.11 |
ENST00000263379.2 |
IL27RA |
interleukin 27 receptor, alpha |
| chr4_-_151936416 | 0.11 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr8_-_81083890 | 0.11 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chr7_+_130126165 | 0.11 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr15_+_65134088 | 0.11 |
ENST00000323544.4 ENST00000437723.1 |
PLEKHO2 AC069368.3 |
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr11_-_89956461 | 0.10 |
ENST00000320585.6 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr7_+_39017504 | 0.10 |
ENST00000403058.1 |
POU6F2 |
POU class 6 homeobox 2 |
| chr4_+_154387480 | 0.10 |
ENST00000409663.3 ENST00000440693.1 ENST00000409959.3 |
KIAA0922 |
KIAA0922 |
| chr13_-_31736478 | 0.10 |
ENST00000445273.2 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr1_-_184006611 | 0.10 |
ENST00000546159.1 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr19_-_49250054 | 0.09 |
ENST00000602105.1 ENST00000332955.2 |
IZUMO1 |
izumo sperm-egg fusion 1 |
| chr13_-_31736027 | 0.09 |
ENST00000380406.5 ENST00000320027.5 ENST00000380405.4 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr13_-_31736132 | 0.09 |
ENST00000429785.2 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr8_-_81083731 | 0.09 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr6_+_32121908 | 0.09 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr7_+_23637763 | 0.09 |
ENST00000410069.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr17_+_56315936 | 0.08 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr12_+_11802753 | 0.08 |
ENST00000396373.4 |
ETV6 |
ets variant 6 |
| chr6_+_32121789 | 0.08 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr14_-_67859422 | 0.08 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr1_+_18958008 | 0.08 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr14_-_102553371 | 0.08 |
ENST00000553585.1 ENST00000216281.8 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr2_-_219151487 | 0.08 |
ENST00000444881.1 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
| chr6_+_32121218 | 0.07 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr4_-_186877806 | 0.07 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_186877502 | 0.07 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_+_42584847 | 0.07 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr3_+_39509070 | 0.07 |
ENST00000354668.4 ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr3_-_183735731 | 0.07 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr17_+_35294075 | 0.07 |
ENST00000254457.5 |
LHX1 |
LIM homeobox 1 |
| chr16_+_71392616 | 0.07 |
ENST00000349553.5 ENST00000302628.4 ENST00000562305.1 |
CALB2 |
calbindin 2 |
| chr6_+_50681541 | 0.07 |
ENST00000008391.3 |
TFAP2D |
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr9_-_97090926 | 0.07 |
ENST00000335456.7 ENST00000253262.4 ENST00000341207.4 |
NUTM2F |
NUT family member 2F |
| chr5_-_159739532 | 0.07 |
ENST00000520748.1 ENST00000393977.3 ENST00000257536.7 |
CCNJL |
cyclin J-like |
| chr2_-_97405775 | 0.07 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr5_+_56469843 | 0.07 |
ENST00000514387.2 |
GPBP1 |
GC-rich promoter binding protein 1 |
| chr7_-_4901625 | 0.06 |
ENST00000404991.1 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
| chr6_+_31795506 | 0.06 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr15_-_81616446 | 0.06 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr12_-_10007448 | 0.06 |
ENST00000538152.1 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr11_-_133826852 | 0.06 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr12_+_52404270 | 0.06 |
ENST00000552049.1 ENST00000546756.1 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr1_-_112531777 | 0.06 |
ENST00000315987.2 ENST00000302127.4 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr15_+_42651691 | 0.06 |
ENST00000357568.3 ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3 |
calpain 3, (p94) |
| chr14_+_103800513 | 0.05 |
ENST00000560338.1 ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chr18_-_812231 | 0.05 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr10_+_135340859 | 0.05 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr5_+_56469939 | 0.05 |
ENST00000506184.2 |
GPBP1 |
GC-rich promoter binding protein 1 |
| chr6_+_42531798 | 0.05 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr20_+_55904815 | 0.05 |
ENST00000371263.3 ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11 |
SPO11 meiotic protein covalently bound to DSB |
| chr5_+_56469775 | 0.05 |
ENST00000424459.3 |
GPBP1 |
GC-rich promoter binding protein 1 |
| chrX_+_115301975 | 0.05 |
ENST00000371906.4 |
AGTR2 |
angiotensin II receptor, type 2 |
| chr3_+_39509163 | 0.05 |
ENST00000436143.2 ENST00000441980.2 ENST00000311042.6 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr20_+_31755934 | 0.05 |
ENST00000354932.5 |
BPIFA2 |
BPI fold containing family A, member 2 |
| chr1_-_184006829 | 0.05 |
ENST00000361927.4 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr6_-_135375986 | 0.05 |
ENST00000525067.1 ENST00000367822.5 ENST00000367837.5 |
HBS1L |
HBS1-like (S. cerevisiae) |
| chr17_+_7608511 | 0.05 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr9_-_34589734 | 0.05 |
ENST00000378980.3 |
CNTFR |
ciliary neurotrophic factor receptor |
| chr3_-_123411191 | 0.04 |
ENST00000354792.5 ENST00000508240.1 |
MYLK |
myosin light chain kinase |
| chr2_+_11674213 | 0.04 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr16_+_55600580 | 0.04 |
ENST00000457326.2 |
CAPNS2 |
calpain, small subunit 2 |
| chr6_-_31782813 | 0.04 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr2_-_74570520 | 0.04 |
ENST00000394019.2 ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr18_+_59000815 | 0.04 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr6_-_53213780 | 0.04 |
ENST00000304434.6 ENST00000370918.4 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
| chrY_-_20935572 | 0.04 |
ENST00000382852.1 ENST00000344884.4 ENST00000304790.3 |
HSFY2 |
heat shock transcription factor, Y linked 2 |
| chr9_-_34589700 | 0.04 |
ENST00000351266.4 |
CNTFR |
ciliary neurotrophic factor receptor |
| chrX_-_110655306 | 0.04 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr7_+_129984630 | 0.04 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr12_+_58138800 | 0.04 |
ENST00000547992.1 ENST00000552816.1 ENST00000547472.1 |
TSPAN31 |
tetraspanin 31 |
| chr3_+_132843652 | 0.04 |
ENST00000508711.1 |
TMEM108 |
transmembrane protein 108 |
| chr12_-_43833515 | 0.04 |
ENST00000549670.1 ENST00000395541.2 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr6_+_31783291 | 0.04 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr17_-_74137374 | 0.04 |
ENST00000322957.6 |
FOXJ1 |
forkhead box J1 |
| chr1_-_102462565 | 0.04 |
ENST00000370103.4 |
OLFM3 |
olfactomedin 3 |
| chr9_-_4666337 | 0.04 |
ENST00000381890.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr19_-_344786 | 0.04 |
ENST00000264819.4 |
MIER2 |
mesoderm induction early response 1, family member 2 |
| chr6_-_123958141 | 0.04 |
ENST00000334268.4 |
TRDN |
triadin |
| chr11_-_122933043 | 0.04 |
ENST00000534624.1 ENST00000453788.2 ENST00000527387.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr12_-_45269430 | 0.04 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr3_-_64673668 | 0.04 |
ENST00000498707.1 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chrY_+_20708557 | 0.03 |
ENST00000307393.2 ENST00000309834.4 ENST00000382856.2 |
HSFY1 |
heat shock transcription factor, Y-linked 1 |
| chrX_+_51075658 | 0.03 |
ENST00000356450.2 |
NUDT10 |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr12_+_69979210 | 0.03 |
ENST00000544368.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr17_-_7297833 | 0.03 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr9_+_841690 | 0.03 |
ENST00000382276.3 |
DMRT1 |
doublesex and mab-3 related transcription factor 1 |
| chrX_-_110655391 | 0.03 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr12_+_58138664 | 0.03 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr12_+_2904102 | 0.03 |
ENST00000001008.4 |
FKBP4 |
FK506 binding protein 4, 59kDa |
| chr11_-_71791726 | 0.03 |
ENST00000393695.3 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr3_-_36781352 | 0.03 |
ENST00000416516.2 |
DCLK3 |
doublecortin-like kinase 3 |
| chr12_+_69979446 | 0.03 |
ENST00000543146.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr11_-_71791518 | 0.03 |
ENST00000537217.1 ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr16_+_68119764 | 0.03 |
ENST00000570212.1 ENST00000562926.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr14_-_57277163 | 0.03 |
ENST00000555006.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr10_+_120116527 | 0.03 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr6_-_123957942 | 0.03 |
ENST00000398178.3 |
TRDN |
triadin |
| chr3_-_49131473 | 0.03 |
ENST00000430979.1 ENST00000357496.2 ENST00000437939.1 |
QRICH1 |
glutamine-rich 1 |
| chr10_-_1246300 | 0.03 |
ENST00000381310.3 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr3_+_96533413 | 0.03 |
ENST00000470610.2 ENST00000389672.5 |
EPHA6 |
EPH receptor A6 |
| chr12_+_69979113 | 0.03 |
ENST00000299300.6 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr4_-_159644507 | 0.03 |
ENST00000307720.3 |
PPID |
peptidylprolyl isomerase D |
| chr10_-_1246317 | 0.03 |
ENST00000381305.1 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr1_-_6453426 | 0.03 |
ENST00000545482.1 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr20_+_4666882 | 0.03 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr1_+_168195229 | 0.03 |
ENST00000271375.4 ENST00000367825.3 |
SFT2D2 |
SFT2 domain containing 2 |
| chr14_+_38091270 | 0.02 |
ENST00000553443.1 |
TTC6 |
tetratricopeptide repeat domain 6 |
| chr3_-_49131013 | 0.02 |
ENST00000424300.1 |
QRICH1 |
glutamine-rich 1 |
| chr18_-_76738843 | 0.02 |
ENST00000575722.1 ENST00000583511.1 |
RP11-849I19.1 |
RP11-849I19.1 |
| chr20_+_18794370 | 0.02 |
ENST00000377428.2 |
SCP2D1 |
SCP2 sterol-binding domain containing 1 |
| chr19_-_49522727 | 0.02 |
ENST00000600007.1 |
CTB-60B18.10 |
CTB-60B18.10 |
| chr2_-_178128250 | 0.02 |
ENST00000448782.1 ENST00000446151.2 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr1_-_6453399 | 0.02 |
ENST00000608083.1 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr21_-_30445886 | 0.02 |
ENST00000431234.1 ENST00000540844.1 ENST00000286788.4 |
CCT8 |
chaperonin containing TCP1, subunit 8 (theta) |
| chr22_+_41253080 | 0.02 |
ENST00000541156.1 ENST00000414396.1 ENST00000357137.4 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr1_+_192544857 | 0.02 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr11_-_111781554 | 0.02 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr14_-_57277178 | 0.02 |
ENST00000339475.5 ENST00000554559.1 ENST00000555804.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr6_-_53213587 | 0.02 |
ENST00000542638.1 ENST00000370913.5 ENST00000541407.1 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
| chr6_+_32709119 | 0.02 |
ENST00000374940.3 |
HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
| chr7_-_26240357 | 0.02 |
ENST00000354667.4 ENST00000356674.7 |
HNRNPA2B1 |
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr7_-_33080506 | 0.02 |
ENST00000381626.2 ENST00000409467.1 ENST00000449201.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr20_+_61867235 | 0.02 |
ENST00000342412.6 ENST00000217169.3 |
BIRC7 |
baculoviral IAP repeat containing 7 |
| chr12_+_107078474 | 0.02 |
ENST00000552866.1 ENST00000229387.5 |
RFX4 |
regulatory factor X, 4 (influences HLA class II expression) |
| chr10_+_18240814 | 0.02 |
ENST00000377374.4 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr6_+_146350267 | 0.02 |
ENST00000355289.4 ENST00000282753.1 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr2_-_85581623 | 0.02 |
ENST00000449375.1 ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT |
retinol saturase (all-trans-retinol 13,14-reductase) |
| chrX_-_30993201 | 0.02 |
ENST00000288422.2 ENST00000378932.2 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr19_-_49560987 | 0.02 |
ENST00000596965.1 |
CGB7 |
chorionic gonadotropin, beta polypeptide 7 |
| chr6_-_30658745 | 0.02 |
ENST00000376420.5 ENST00000376421.5 |
NRM |
nurim (nuclear envelope membrane protein) |
| chr10_-_90611566 | 0.02 |
ENST00000371930.4 |
ANKRD22 |
ankyrin repeat domain 22 |
| chr4_+_123161120 | 0.02 |
ENST00000446180.1 |
KIAA1109 |
KIAA1109 |
| chr19_+_6135646 | 0.02 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chrX_-_101718085 | 0.02 |
ENST00000372750.1 ENST00000372752.1 |
NXF2B |
nuclear RNA export factor 2B |
| chr15_-_58357866 | 0.01 |
ENST00000537372.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr12_-_123011536 | 0.01 |
ENST00000331738.7 ENST00000354654.2 |
RSRC2 |
arginine/serine-rich coiled-coil 2 |
| chr2_+_219824357 | 0.01 |
ENST00000302625.4 |
CDK5R2 |
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr12_-_45270077 | 0.01 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr14_-_25479811 | 0.01 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chrX_-_72347916 | 0.01 |
ENST00000373518.1 |
NAP1L6 |
nucleosome assembly protein 1-like 6 |
| chrX_+_101478829 | 0.01 |
ENST00000372763.1 ENST00000372758.1 |
NXF2 |
nuclear RNA export factor 2 |
| chr2_-_85581701 | 0.01 |
ENST00000295802.4 |
RETSAT |
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr11_-_111781610 | 0.01 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_122932730 | 0.01 |
ENST00000532182.1 ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr6_-_32339649 | 0.01 |
ENST00000527965.1 ENST00000532023.1 ENST00000447241.2 ENST00000375007.4 ENST00000534588.1 |
C6orf10 |
chromosome 6 open reading frame 10 |
| chr5_-_135701164 | 0.01 |
ENST00000355180.3 ENST00000426057.2 ENST00000513104.1 |
TRPC7 |
transient receptor potential cation channel, subfamily C, member 7 |
| chr7_+_148936732 | 0.01 |
ENST00000335870.2 |
ZNF212 |
zinc finger protein 212 |
| chr10_+_18240753 | 0.01 |
ENST00000377369.2 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr16_-_30457048 | 0.01 |
ENST00000500504.2 ENST00000542752.1 |
SEPHS2 |
selenophosphate synthetase 2 |
| chr2_+_202937972 | 0.01 |
ENST00000541917.1 ENST00000295844.3 |
AC079354.1 |
uncharacterized protein KIAA2012 |
| chr6_-_32339671 | 0.01 |
ENST00000442822.2 ENST00000375015.4 ENST00000533191.1 |
C6orf10 |
chromosome 6 open reading frame 10 |
| chr2_+_158733088 | 0.01 |
ENST00000605860.1 |
UPP2 |
uridine phosphorylase 2 |
| chr17_-_71410794 | 0.01 |
ENST00000424778.1 |
SDK2 |
sidekick cell adhesion molecule 2 |
| chr7_-_36764142 | 0.01 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr7_-_14880892 | 0.01 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr11_-_65769594 | 0.01 |
ENST00000532707.1 ENST00000533544.1 ENST00000526451.1 ENST00000312234.2 ENST00000530462.1 ENST00000525767.1 ENST00000529964.1 ENST00000527249.1 |
EIF1AD |
eukaryotic translation initiation factor 1A domain containing |
| chr17_+_41052808 | 0.01 |
ENST00000592383.1 ENST00000253801.2 ENST00000585489.1 |
G6PC |
glucose-6-phosphatase, catalytic subunit |
| chr6_+_41303372 | 0.00 |
ENST00000373083.4 ENST00000373089.5 ENST00000373086.3 |
NCR2 |
natural cytotoxicity triggering receptor 2 |
| chr16_+_226658 | 0.00 |
ENST00000320868.5 ENST00000397797.1 |
HBA1 |
hemoglobin, alpha 1 |
| chr19_+_782755 | 0.00 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr6_-_155635583 | 0.00 |
ENST00000367166.4 |
TFB1M |
transcription factor B1, mitochondrial |
| chr19_+_54606145 | 0.00 |
ENST00000485876.1 ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr8_+_87878640 | 0.00 |
ENST00000518476.1 |
CNBD1 |
cyclic nucleotide binding domain containing 1 |
| chr12_-_123011476 | 0.00 |
ENST00000528279.1 ENST00000344591.4 ENST00000526560.2 |
RSRC2 |
arginine/serine-rich coiled-coil 2 |
| chr5_-_35089722 | 0.00 |
ENST00000511486.1 ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR |
prolactin receptor |
| chr1_-_216596738 | 0.00 |
ENST00000307340.3 ENST00000366943.2 ENST00000366942.3 |
USH2A |
Usher syndrome 2A (autosomal recessive, mild) |
| chr7_+_75931861 | 0.00 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr15_+_78556809 | 0.00 |
ENST00000343789.3 ENST00000394852.3 |
DNAJA4 |
DnaJ (Hsp40) homolog, subfamily A, member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.3 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.2 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0060067 | cervix development(GO:0060067) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |