Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for IRF7
Z-value: 2.19
Transcription factors associated with IRF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF7
|
ENSG00000185507.15 | IRF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF7 | hg19_v2_chr11_-_615570_615728 | 0.49 | 2.2e-01 | Click! |
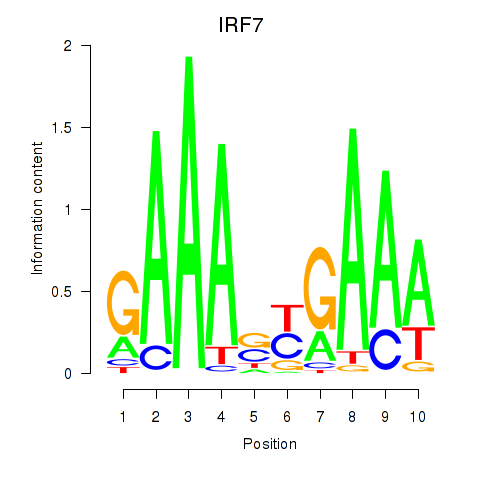
Activity profile of IRF7 motif
Sorted Z-values of IRF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 5.97 |
ENST00000547937.1 |
DCN |
decorin |
| chr13_+_102104980 | 3.96 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_+_97597148 | 3.65 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr12_-_91572278 | 3.07 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr9_-_95186739 | 2.90 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr5_+_92919043 | 2.71 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr12_-_91576750 | 2.62 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr12_-_91576561 | 2.51 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr1_+_155829286 | 2.44 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr1_+_79086088 | 2.43 |
ENST00000370751.5 ENST00000342282.3 |
IFI44L |
interferon-induced protein 44-like |
| chr13_-_43566301 | 2.24 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr1_-_85870177 | 2.12 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_91573132 | 2.08 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573316 | 2.00 |
ENST00000393155.1 |
DCN |
decorin |
| chr1_+_221051699 | 1.95 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr2_-_188419200 | 1.94 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr4_-_138453606 | 1.86 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr8_+_17434689 | 1.86 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr11_-_2160180 | 1.82 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr2_-_190044480 | 1.82 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chrX_+_102883620 | 1.82 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr1_+_162602244 | 1.79 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr2_-_145277640 | 1.78 |
ENST00000539609.3 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr4_-_70626314 | 1.75 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr2_-_188419078 | 1.68 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_+_221054584 | 1.58 |
ENST00000549319.1 |
HLX |
H2.0-like homeobox |
| chr2_-_145277569 | 1.56 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr11_+_19799327 | 1.54 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr5_-_149535421 | 1.49 |
ENST00000261799.4 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
| chr2_+_210444748 | 1.43 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr8_+_77593448 | 1.42 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_+_77593474 | 1.37 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr1_+_79115503 | 1.32 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr5_+_32710736 | 1.30 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr6_+_72922590 | 1.25 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr2_+_210444142 | 1.25 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr6_+_72922505 | 1.24 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_-_114035026 | 1.22 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr13_-_33780133 | 1.22 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr3_-_114477787 | 1.22 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_156696362 | 1.20 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_140734570 | 1.19 |
ENST00000571252.1 |
PCDHGA4 |
protocadherin gamma subfamily A, 4 |
| chr9_+_90112741 | 1.18 |
ENST00000469640.2 |
DAPK1 |
death-associated protein kinase 1 |
| chr16_+_86600857 | 1.17 |
ENST00000320354.4 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr11_-_119293872 | 1.15 |
ENST00000524970.1 |
THY1 |
Thy-1 cell surface antigen |
| chr9_-_20622478 | 1.14 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chrX_+_80457442 | 1.14 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr3_+_154797877 | 1.13 |
ENST00000462745.1 ENST00000493237.1 |
MME |
membrane metallo-endopeptidase |
| chr9_+_90112767 | 1.13 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr4_-_39640513 | 1.09 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr3_+_141103634 | 1.04 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_-_67138015 | 1.03 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr1_-_150693318 | 1.01 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr7_-_122526799 | 1.01 |
ENST00000334010.7 ENST00000313070.7 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chrX_+_22050546 | 1.01 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr10_+_91092241 | 1.00 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr12_+_15699286 | 1.00 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr11_-_57335280 | 0.98 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr9_+_4985016 | 0.95 |
ENST00000539801.1 |
JAK2 |
Janus kinase 2 |
| chr16_-_67970990 | 0.94 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chrX_+_102883887 | 0.93 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr4_-_138453559 | 0.91 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr10_+_91152303 | 0.91 |
ENST00000371804.3 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 |
| chr10_-_92681033 | 0.90 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr6_-_154751629 | 0.89 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr7_-_92777606 | 0.89 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr6_-_87804815 | 0.88 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr1_+_197881592 | 0.87 |
ENST00000367391.1 ENST00000367390.3 |
LHX9 |
LIM homeobox 9 |
| chr11_-_111794446 | 0.87 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr13_-_33112823 | 0.87 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr2_-_202562774 | 0.86 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr2_-_152118352 | 0.84 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr5_+_140749803 | 0.84 |
ENST00000576222.1 |
PCDHGB3 |
protocadherin gamma subfamily B, 3 |
| chr3_-_112329110 | 0.83 |
ENST00000479368.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chrX_-_34675391 | 0.82 |
ENST00000275954.3 |
TMEM47 |
transmembrane protein 47 |
| chr1_+_150245177 | 0.81 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr11_+_77532155 | 0.80 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr2_+_102721023 | 0.80 |
ENST00000409589.1 ENST00000409329.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr4_+_113739244 | 0.80 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr9_-_21305312 | 0.80 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr5_-_95158644 | 0.79 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr14_+_76044934 | 0.79 |
ENST00000238667.4 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr4_+_142557717 | 0.78 |
ENST00000320650.4 ENST00000296545.7 |
IL15 |
interleukin 15 |
| chrX_+_51486481 | 0.78 |
ENST00000340438.4 |
GSPT2 |
G1 to S phase transition 2 |
| chr11_+_77532233 | 0.77 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr11_+_63304273 | 0.76 |
ENST00000439013.2 ENST00000255688.3 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
| chr1_+_210406121 | 0.76 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr13_+_32838801 | 0.76 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr6_-_167571817 | 0.75 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr2_-_158345462 | 0.75 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr9_-_123812542 | 0.74 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr4_+_142557771 | 0.73 |
ENST00000514653.1 |
IL15 |
interleukin 15 |
| chr6_-_30709980 | 0.72 |
ENST00000416018.1 ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1 |
flotillin 1 |
| chr10_+_91174314 | 0.72 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chrX_+_103031421 | 0.72 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr12_-_76879852 | 0.71 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chrX_+_9431324 | 0.69 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr22_-_32651326 | 0.69 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr3_+_29322437 | 0.69 |
ENST00000434693.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr3_-_49851313 | 0.69 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr2_-_231084617 | 0.69 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr2_+_27301435 | 0.69 |
ENST00000380320.4 |
EMILIN1 |
elastin microfibril interfacer 1 |
| chr6_-_30710265 | 0.68 |
ENST00000438162.1 ENST00000454845.1 |
FLOT1 |
flotillin 1 |
| chr5_-_124080203 | 0.68 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr3_+_141105705 | 0.67 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr21_+_17909594 | 0.67 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chrX_+_102631248 | 0.67 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr3_+_187086120 | 0.66 |
ENST00000259030.2 |
RTP4 |
receptor (chemosensory) transporter protein 4 |
| chr11_-_96076334 | 0.66 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr6_-_24936170 | 0.66 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr19_+_39786962 | 0.65 |
ENST00000333625.2 |
IFNL1 |
interferon, lambda 1 |
| chr10_+_91087651 | 0.65 |
ENST00000371818.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chrX_+_103031758 | 0.65 |
ENST00000303958.2 ENST00000361621.2 |
PLP1 |
proteolipid protein 1 |
| chr7_-_80141328 | 0.65 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr1_+_78470530 | 0.65 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr14_-_35182994 | 0.65 |
ENST00000341223.3 |
CFL2 |
cofilin 2 (muscle) |
| chr10_+_92980517 | 0.64 |
ENST00000336126.5 |
PCGF5 |
polycomb group ring finger 5 |
| chr6_+_101846664 | 0.63 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr12_+_117176113 | 0.63 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr3_-_114477962 | 0.61 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_-_202562716 | 0.61 |
ENST00000428900.2 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr9_-_21202204 | 0.60 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr1_+_948803 | 0.60 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr2_-_37899323 | 0.60 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_+_94542459 | 0.60 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr2_+_166095898 | 0.59 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr7_-_14029515 | 0.59 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chrX_+_22056165 | 0.59 |
ENST00000535894.1 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_150245099 | 0.59 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr12_+_32260085 | 0.59 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr17_+_41158742 | 0.58 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chr2_+_201980961 | 0.58 |
ENST00000342795.5 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr17_-_33864772 | 0.58 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr3_+_29322803 | 0.58 |
ENST00000396583.3 ENST00000383767.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr6_+_122931366 | 0.57 |
ENST00000368452.2 ENST00000368448.1 ENST00000392490.1 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr9_-_95298314 | 0.57 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr1_-_104239076 | 0.56 |
ENST00000370080.3 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr5_+_140625147 | 0.56 |
ENST00000231173.3 |
PCDHB15 |
protocadherin beta 15 |
| chr2_+_201980827 | 0.56 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_+_172950227 | 0.56 |
ENST00000341900.6 |
DLX1 |
distal-less homeobox 1 |
| chr6_+_147527103 | 0.55 |
ENST00000179882.6 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
| chr19_+_49977466 | 0.55 |
ENST00000596435.1 ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr12_+_79258547 | 0.55 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr8_-_116680833 | 0.55 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr1_-_193155729 | 0.55 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr14_-_35183755 | 0.54 |
ENST00000555765.1 |
CFL2 |
cofilin 2 (muscle) |
| chr3_+_141105235 | 0.54 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr1_-_149900122 | 0.54 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr8_+_79428539 | 0.53 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chrX_+_77166172 | 0.53 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr8_-_93107696 | 0.53 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_+_38586068 | 0.53 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr6_-_30710510 | 0.52 |
ENST00000376389.3 |
FLOT1 |
flotillin 1 |
| chr8_-_93029520 | 0.52 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_49977818 | 0.51 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr9_-_21077939 | 0.50 |
ENST00000380232.2 |
IFNB1 |
interferon, beta 1, fibroblast |
| chr10_+_94608245 | 0.50 |
ENST00000443748.2 ENST00000260762.6 |
EXOC6 |
exocyst complex component 6 |
| chr21_+_27011584 | 0.50 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr14_-_70883708 | 0.50 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr1_-_227505289 | 0.49 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_-_63005405 | 0.49 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr14_+_24605389 | 0.48 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr8_-_93107827 | 0.48 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_-_37393406 | 0.48 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr11_+_20044096 | 0.47 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr10_-_70092671 | 0.47 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr15_+_96875657 | 0.47 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_12717892 | 0.47 |
ENST00000379350.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr10_+_94590910 | 0.47 |
ENST00000371547.4 |
EXOC6 |
exocyst complex component 6 |
| chr4_+_186990298 | 0.46 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr6_-_15586238 | 0.46 |
ENST00000462989.2 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr6_-_36515177 | 0.46 |
ENST00000229812.7 |
STK38 |
serine/threonine kinase 38 |
| chr5_+_139505520 | 0.46 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chrX_+_153770421 | 0.46 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr6_-_30710447 | 0.46 |
ENST00000456573.2 |
FLOT1 |
flotillin 1 |
| chr19_+_39759154 | 0.45 |
ENST00000331982.5 |
IFNL2 |
interferon, lambda 2 |
| chr4_-_77134742 | 0.44 |
ENST00000452464.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr10_+_91061712 | 0.44 |
ENST00000371826.3 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
| chr1_+_210502238 | 0.44 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr2_-_231084659 | 0.44 |
ENST00000258381.6 ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110 |
SP110 nuclear body protein |
| chr10_+_85954377 | 0.44 |
ENST00000332904.3 ENST00000372117.3 |
CDHR1 |
cadherin-related family member 1 |
| chr17_-_33759509 | 0.44 |
ENST00000304905.5 |
SLFN12 |
schlafen family member 12 |
| chr2_+_201981527 | 0.44 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr1_-_114301503 | 0.43 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr6_+_151561085 | 0.43 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr3_-_47950745 | 0.43 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr12_-_92536433 | 0.42 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr19_+_30863271 | 0.42 |
ENST00000355537.3 |
ZNF536 |
zinc finger protein 536 |
| chr6_+_10585979 | 0.42 |
ENST00000265012.4 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr7_+_134331550 | 0.42 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr1_-_243326612 | 0.41 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr6_-_85473156 | 0.41 |
ENST00000606784.1 ENST00000606325.1 |
TBX18 |
T-box 18 |
| chrX_-_71525742 | 0.41 |
ENST00000450875.1 ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chrX_+_100878079 | 0.40 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr3_+_29323043 | 0.40 |
ENST00000452462.1 ENST00000456853.1 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr10_+_115439630 | 0.40 |
ENST00000369318.3 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr3_+_185046676 | 0.39 |
ENST00000428617.1 ENST00000443863.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr7_-_134143841 | 0.39 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr13_+_112721913 | 0.39 |
ENST00000330949.1 |
SOX1 |
SRY (sex determining region Y)-box 1 |
| chr1_-_182361327 | 0.39 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 2.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.6 | 1.9 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.6 | 3.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.6 | 1.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 3.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 4.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 1.6 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.4 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 3.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 1.2 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.4 | 1.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 1.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.4 | 1.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.3 | 1.0 | GO:0051598 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.3 | 1.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 0.9 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 0.3 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.3 | 0.8 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 2.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 2.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.2 | 2.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.8 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.2 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.0 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 0.6 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.2 | 0.9 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 0.6 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.5 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 1.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 2.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.9 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 0.6 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 2.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.9 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 1.0 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 1.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 2.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.4 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 1.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:0006059 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.7 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.4 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 | 0.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.5 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 2.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.9 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.3 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.1 | 0.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.3 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 1.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.6 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 1.5 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.8 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.6 | GO:0051967 | regulation of short-term neuronal synaptic plasticity(GO:0048172) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 1.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 2.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.6 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 2.8 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.3 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.7 | 2.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 2.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 1.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 1.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.6 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.2 | 2.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 3.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 2.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.4 | GO:0031094 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.9 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 0.3 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 2.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 8.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 3.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 2.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 0.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.7 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 18.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.5 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 0.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.5 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.2 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.6 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 2.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 2.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 4.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 3.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0019960 | C-X3-C chemokine receptor activity(GO:0016495) C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 21.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 4.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 18.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 3.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |