Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for IRF9
Z-value: 1.13
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
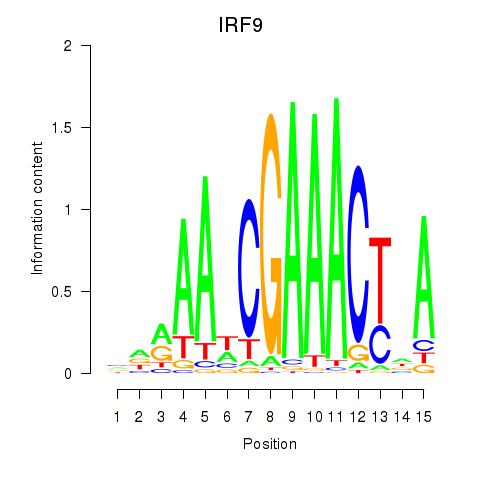
IRF9
|
ENSG00000213928.4 | IRF9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | 0.72 | 4.5e-02 | Click! |
Activity profile of IRF9 motif
Sorted Z-values of IRF9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79086088 | 2.54 |
ENST00000370751.5 ENST00000342282.3 |
IFI44L |
interferon-induced protein 44-like |
| chr6_+_72922590 | 1.16 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr6_+_72922505 | 1.16 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr1_+_162602244 | 1.00 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr18_-_25616519 | 0.94 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr13_-_43566301 | 0.94 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr17_+_6659153 | 0.82 |
ENST00000441631.1 ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1 |
XIAP associated factor 1 |
| chr1_+_79115503 | 0.79 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr5_+_32710736 | 0.65 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_-_57335280 | 0.60 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr14_+_24630465 | 0.52 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr3_-_122283079 | 0.52 |
ENST00000471785.1 ENST00000466126.1 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr7_+_134551583 | 0.51 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr3_-_122283424 | 0.49 |
ENST00000477522.2 ENST00000360356.2 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr10_-_25241499 | 0.48 |
ENST00000376378.1 ENST00000376376.3 ENST00000320152.6 |
PRTFDC1 |
phosphoribosyl transferase domain containing 1 |
| chrX_-_63005405 | 0.47 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_-_86383157 | 0.44 |
ENST00000393324.3 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr6_-_82462425 | 0.43 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr11_-_86383650 | 0.41 |
ENST00000526944.1 ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr10_+_91092241 | 0.38 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_+_948803 | 0.37 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr6_-_24936170 | 0.37 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr15_+_91643442 | 0.36 |
ENST00000394232.1 |
SV2B |
synaptic vesicle glycoprotein 2B |
| chrX_+_22056165 | 0.36 |
ENST00000535894.1 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr10_+_91174314 | 0.33 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chr2_-_152146385 | 0.33 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr4_+_186990298 | 0.32 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr17_+_78234625 | 0.31 |
ENST00000508628.2 ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213 |
ring finger protein 213 |
| chr19_-_17516449 | 0.31 |
ENST00000252593.6 |
BST2 |
bone marrow stromal cell antigen 2 |
| chr17_+_46908350 | 0.27 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr4_-_77134742 | 0.27 |
ENST00000452464.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr3_+_151986709 | 0.27 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr19_+_10196981 | 0.26 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr11_-_615570 | 0.26 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr10_+_115439630 | 0.25 |
ENST00000369318.3 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr16_-_15736881 | 0.25 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr7_+_100728720 | 0.25 |
ENST00000306085.6 ENST00000412507.1 |
TRIM56 |
tripartite motif containing 56 |
| chr16_+_28875126 | 0.25 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr3_+_187086120 | 0.24 |
ENST00000259030.2 |
RTP4 |
receptor (chemosensory) transporter protein 4 |
| chr10_+_115439282 | 0.23 |
ENST00000369321.2 ENST00000345633.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr14_+_100531615 | 0.23 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr6_+_6588902 | 0.23 |
ENST00000230568.4 |
LY86 |
lymphocyte antigen 86 |
| chr3_-_121379739 | 0.23 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr10_+_115439699 | 0.22 |
ENST00000369315.1 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr4_-_77135046 | 0.22 |
ENST00000264896.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr3_-_146262365 | 0.19 |
ENST00000448787.2 |
PLSCR1 |
phospholipid scramblase 1 |
| chr17_-_4167142 | 0.19 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr3_-_146262352 | 0.18 |
ENST00000462666.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chrX_-_39956656 | 0.17 |
ENST00000397354.3 ENST00000378444.4 |
BCOR |
BCL6 corepressor |
| chrX_-_48858630 | 0.17 |
ENST00000376425.3 ENST00000376444.3 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr3_-_146262428 | 0.17 |
ENST00000486631.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr6_+_126240442 | 0.17 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chrX_-_48858667 | 0.16 |
ENST00000376423.4 ENST00000376441.1 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr2_-_152118352 | 0.16 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr9_-_21995300 | 0.16 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr1_+_241695424 | 0.16 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr13_+_50070491 | 0.15 |
ENST00000496612.1 ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11 |
PHD finger protein 11 |
| chr19_+_17516909 | 0.15 |
ENST00000601007.1 ENST00000594913.1 ENST00000599975.1 ENST00000600514.1 |
CTD-2521M24.9 MVB12A |
CTD-2521M24.9 multivesicular body subunit 12A |
| chr3_-_28390581 | 0.15 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr14_-_67981870 | 0.15 |
ENST00000555994.1 |
TMEM229B |
transmembrane protein 229B |
| chr3_-_146262488 | 0.14 |
ENST00000487389.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr1_+_236687881 | 0.14 |
ENST00000526634.1 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr10_+_91061712 | 0.14 |
ENST00000371826.3 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
| chr1_+_154377669 | 0.14 |
ENST00000368485.3 ENST00000344086.4 |
IL6R |
interleukin 6 receptor |
| chr13_+_50070077 | 0.14 |
ENST00000378319.3 ENST00000426879.1 |
PHF11 |
PHD finger protein 11 |
| chr9_-_21995249 | 0.14 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr2_+_205410516 | 0.13 |
ENST00000406610.2 ENST00000462231.1 |
PARD3B |
par-3 family cell polarity regulator beta |
| chr3_-_146262293 | 0.13 |
ENST00000448205.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr15_+_74287035 | 0.13 |
ENST00000395132.2 ENST00000268059.6 ENST00000354026.6 ENST00000268058.3 ENST00000565898.1 ENST00000569477.1 ENST00000569965.1 ENST00000567543.1 ENST00000436891.3 ENST00000435786.2 ENST00000564428.1 ENST00000359928.4 |
PML |
promyelocytic leukemia |
| chr12_-_121476750 | 0.13 |
ENST00000543677.1 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121477039 | 0.12 |
ENST00000257570.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr4_+_37892682 | 0.12 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr11_-_67141090 | 0.12 |
ENST00000312438.7 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
| chr1_-_207119738 | 0.12 |
ENST00000356495.4 |
PIGR |
polymeric immunoglobulin receptor |
| chr22_+_18632666 | 0.12 |
ENST00000215794.7 |
USP18 |
ubiquitin specific peptidase 18 |
| chr3_-_146262637 | 0.12 |
ENST00000472349.1 ENST00000342435.4 |
PLSCR1 |
phospholipid scramblase 1 |
| chr20_+_11898507 | 0.12 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr12_+_6881678 | 0.12 |
ENST00000441671.2 ENST00000203629.2 |
LAG3 |
lymphocyte-activation gene 3 |
| chr12_-_56753858 | 0.11 |
ENST00000314128.4 ENST00000557235.1 ENST00000418572.2 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
| chr6_-_110736742 | 0.11 |
ENST00000368924.3 ENST00000368923.3 |
DDO |
D-aspartate oxidase |
| chr12_-_27167233 | 0.11 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
| chr1_+_241695670 | 0.10 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_+_17516531 | 0.10 |
ENST00000528911.1 ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A CTD-2521M24.9 |
multivesicular body subunit 12A CTD-2521M24.9 |
| chr7_+_18535893 | 0.09 |
ENST00000432645.2 ENST00000441542.2 |
HDAC9 |
histone deacetylase 9 |
| chr1_-_182558374 | 0.09 |
ENST00000367559.3 ENST00000539397.1 |
RNASEL |
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
| chr5_-_142783175 | 0.08 |
ENST00000231509.3 ENST00000394464.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_38296576 | 0.08 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr14_-_67981916 | 0.08 |
ENST00000357461.2 |
TMEM229B |
transmembrane protein 229B |
| chr3_+_122399444 | 0.08 |
ENST00000474629.2 |
PARP14 |
poly (ADP-ribose) polymerase family, member 14 |
| chr11_-_615942 | 0.08 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr2_-_37384175 | 0.07 |
ENST00000411537.2 ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2 |
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr5_-_142783694 | 0.07 |
ENST00000394466.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_-_105584560 | 0.07 |
ENST00000336775.5 |
BVES |
blood vessel epicardial substance |
| chr3_-_122283100 | 0.06 |
ENST00000492382.1 ENST00000462315.1 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_28390415 | 0.06 |
ENST00000414162.1 ENST00000420543.2 |
AZI2 |
5-azacytidine induced 2 |
| chr3_-_28390298 | 0.06 |
ENST00000457172.1 |
AZI2 |
5-azacytidine induced 2 |
| chr3_-_141747459 | 0.06 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr19_+_16254488 | 0.05 |
ENST00000588246.1 ENST00000593031.1 |
HSH2D |
hematopoietic SH2 domain containing |
| chr10_-_7513904 | 0.05 |
ENST00000420395.1 |
RP5-1031D4.2 |
RP5-1031D4.2 |
| chr12_-_121476959 | 0.04 |
ENST00000339275.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr20_+_61436146 | 0.04 |
ENST00000290291.6 |
OGFR |
opioid growth factor receptor |
| chr9_+_102668915 | 0.03 |
ENST00000259400.6 ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17 |
syntaxin 17 |
| chr9_-_33264557 | 0.03 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr2_+_205410723 | 0.03 |
ENST00000358768.2 ENST00000351153.1 ENST00000349953.3 |
PARD3B |
par-3 family cell polarity regulator beta |
| chr8_-_27941380 | 0.02 |
ENST00000413272.2 ENST00000341513.6 |
NUGGC |
nuclear GTPase, germinal center associated |
| chr18_+_3262954 | 0.02 |
ENST00000584539.1 |
MYL12B |
myosin, light chain 12B, regulatory |
| chr6_-_33282024 | 0.02 |
ENST00000475304.1 ENST00000489157.1 |
TAPBP |
TAP binding protein (tapasin) |
| chr3_-_142166846 | 0.01 |
ENST00000463916.1 ENST00000544157.1 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr16_+_28962128 | 0.01 |
ENST00000564978.1 ENST00000320805.4 |
NFATC2IP |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein |
| chr3_-_142166904 | 0.01 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr6_-_33281979 | 0.01 |
ENST00000426633.2 ENST00000467025.1 |
TAPBP |
TAP binding protein (tapasin) |
| chr18_-_67624160 | 0.01 |
ENST00000581982.1 ENST00000280200.4 |
CD226 |
CD226 molecule |
| chr3_+_122283064 | 0.00 |
ENST00000296161.4 |
DTX3L |
deltex 3-like (Drosophila) |
| chr9_+_5510558 | 0.00 |
ENST00000397747.3 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.9 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 2.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.5 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.6 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.3 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.9 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.4 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.7 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 2.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.9 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |