Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
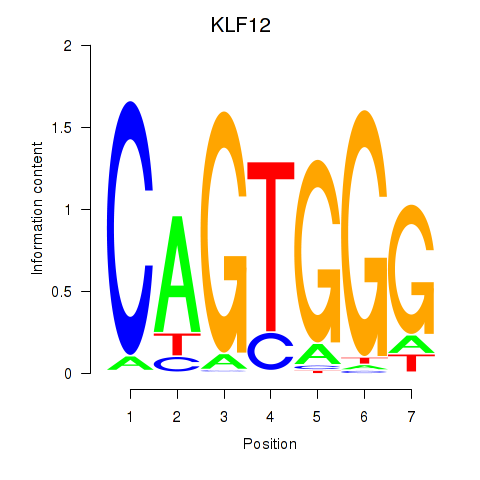
Results for KLF12
Z-value: 0.05
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | KLF12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | 0.02 | 9.7e-01 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153750622 | 0.75 |
ENST00000532853.1 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr11_-_102401469 | 0.43 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_+_109792641 | 0.36 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr17_+_1665253 | 0.32 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr3_-_49722523 | 0.30 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr2_+_128180842 | 0.28 |
ENST00000402125.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr11_+_117070037 | 0.28 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr12_-_54778471 | 0.28 |
ENST00000550120.1 ENST00000394313.2 ENST00000547210.1 |
ZNF385A |
zinc finger protein 385A |
| chr17_+_1665345 | 0.27 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr6_+_72596406 | 0.25 |
ENST00000491071.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr5_-_81046922 | 0.24 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr15_+_74218787 | 0.24 |
ENST00000261921.7 |
LOXL1 |
lysyl oxidase-like 1 |
| chr5_-_81046841 | 0.24 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr5_+_145718587 | 0.23 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chr3_+_193853927 | 0.23 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr1_-_46598371 | 0.23 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr5_-_168728103 | 0.23 |
ENST00000519560.1 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr16_-_67978016 | 0.23 |
ENST00000264005.5 |
LCAT |
lecithin-cholesterol acyltransferase |
| chr5_-_81046904 | 0.22 |
ENST00000515395.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr3_-_61237050 | 0.21 |
ENST00000476844.1 ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT |
fragile histidine triad |
| chrX_+_150866779 | 0.20 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr1_+_214161854 | 0.20 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr13_-_67802549 | 0.19 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr13_-_67804445 | 0.18 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr17_-_36831156 | 0.18 |
ENST00000325814.5 |
C17orf96 |
chromosome 17 open reading frame 96 |
| chr1_+_27561007 | 0.18 |
ENST00000319394.3 |
WDTC1 |
WD and tetratricopeptide repeats 1 |
| chr22_-_38699003 | 0.18 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chr6_+_31895467 | 0.18 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_+_27561104 | 0.18 |
ENST00000361771.3 |
WDTC1 |
WD and tetratricopeptide repeats 1 |
| chr11_+_121461097 | 0.17 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr11_-_568369 | 0.17 |
ENST00000534540.1 ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG |
MIR210 host gene (non-protein coding) |
| chr1_-_46598284 | 0.17 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr2_+_203499901 | 0.17 |
ENST00000303116.6 ENST00000392238.2 |
FAM117B |
family with sequence similarity 117, member B |
| chrX_-_99986494 | 0.16 |
ENST00000372989.1 ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4 |
synaptotagmin-like 4 |
| chr12_-_6772303 | 0.16 |
ENST00000396807.4 ENST00000446105.2 ENST00000341550.4 |
ING4 |
inhibitor of growth family, member 4 |
| chr2_+_85646054 | 0.16 |
ENST00000389938.2 |
SH2D6 |
SH2 domain containing 6 |
| chr8_+_123793633 | 0.16 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr2_+_234104079 | 0.16 |
ENST00000417661.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr17_+_7608511 | 0.16 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr9_-_114557207 | 0.15 |
ENST00000374287.3 ENST00000374283.5 |
C9orf84 |
chromosome 9 open reading frame 84 |
| chr11_-_32457075 | 0.15 |
ENST00000448076.3 |
WT1 |
Wilms tumor 1 |
| chr10_-_75401500 | 0.15 |
ENST00000359322.4 |
MYOZ1 |
myozenin 1 |
| chr3_-_48057890 | 0.15 |
ENST00000434267.1 |
MAP4 |
microtubule-associated protein 4 |
| chr16_+_85646891 | 0.14 |
ENST00000393243.1 |
GSE1 |
Gse1 coiled-coil protein |
| chr4_-_20985632 | 0.14 |
ENST00000359001.5 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr20_-_31071309 | 0.14 |
ENST00000326071.4 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr9_+_131084846 | 0.13 |
ENST00000608951.1 |
COQ4 |
coenzyme Q4 |
| chr10_-_91403625 | 0.13 |
ENST00000322191.6 ENST00000342512.3 ENST00000371774.2 |
PANK1 |
pantothenate kinase 1 |
| chr8_+_22022800 | 0.13 |
ENST00000397814.3 |
BMP1 |
bone morphogenetic protein 1 |
| chr15_-_52944231 | 0.13 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr20_+_34287364 | 0.13 |
ENST00000374072.1 ENST00000397416.1 ENST00000336695.4 |
ROMO1 |
reactive oxygen species modulator 1 |
| chr11_-_66360548 | 0.13 |
ENST00000333861.3 |
CCDC87 |
coiled-coil domain containing 87 |
| chr15_+_84115868 | 0.13 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr16_+_85646763 | 0.13 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr4_-_5710257 | 0.13 |
ENST00000344938.1 ENST00000344408.5 |
EVC2 |
Ellis van Creveld syndrome 2 |
| chr1_+_50574585 | 0.12 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr16_+_28889801 | 0.12 |
ENST00000395503.4 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr14_+_102276192 | 0.12 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chrX_-_112084043 | 0.12 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr20_-_31071239 | 0.12 |
ENST00000359676.5 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr20_+_34287194 | 0.12 |
ENST00000374078.1 ENST00000374077.3 |
ROMO1 |
reactive oxygen species modulator 1 |
| chr4_-_170947522 | 0.12 |
ENST00000361618.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr14_+_74417192 | 0.12 |
ENST00000554320.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr1_+_6845384 | 0.12 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr3_-_133614421 | 0.12 |
ENST00000543906.1 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr16_+_28889703 | 0.12 |
ENST00000357084.3 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr10_-_115933942 | 0.12 |
ENST00000369285.3 ENST00000369287.3 ENST00000369286.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr1_-_76398077 | 0.11 |
ENST00000284142.6 |
ASB17 |
ankyrin repeat and SOCS box containing 17 |
| chr2_+_169659121 | 0.11 |
ENST00000397206.2 ENST00000397209.2 ENST00000421711.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chr3_-_133614597 | 0.11 |
ENST00000285208.4 ENST00000460865.3 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr1_-_155177677 | 0.11 |
ENST00000368378.3 ENST00000541990.1 ENST00000457183.2 |
THBS3 |
thrombospondin 3 |
| chr7_-_79082867 | 0.11 |
ENST00000419488.1 ENST00000354212.4 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_+_169658928 | 0.11 |
ENST00000317647.7 ENST00000445023.2 |
NOSTRIN |
nitric oxide synthase trafficking |
| chr1_+_6845497 | 0.10 |
ENST00000473578.1 ENST00000557126.1 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr1_-_6259641 | 0.10 |
ENST00000234875.4 |
RPL22 |
ribosomal protein L22 |
| chr16_-_20911641 | 0.10 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr17_-_76899275 | 0.10 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr6_-_27775694 | 0.10 |
ENST00000377401.2 |
HIST1H2BL |
histone cluster 1, H2bl |
| chr17_-_72864739 | 0.10 |
ENST00000579893.1 ENST00000544854.1 |
FDXR |
ferredoxin reductase |
| chr11_-_72492878 | 0.10 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr12_+_79258547 | 0.10 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr3_-_116163830 | 0.10 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr22_+_47016277 | 0.10 |
ENST00000406902.1 |
GRAMD4 |
GRAM domain containing 4 |
| chr6_-_111804393 | 0.10 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr11_+_66360665 | 0.10 |
ENST00000310190.4 |
CCS |
copper chaperone for superoxide dismutase |
| chr8_+_75736761 | 0.10 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr2_+_220306745 | 0.10 |
ENST00000431523.1 ENST00000396698.1 ENST00000396695.2 |
SPEG |
SPEG complex locus |
| chrX_-_74743080 | 0.10 |
ENST00000373367.3 |
ZDHHC15 |
zinc finger, DHHC-type containing 15 |
| chr2_+_24272543 | 0.10 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr2_-_225266743 | 0.10 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr20_+_34713312 | 0.10 |
ENST00000373946.3 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chrX_-_102531717 | 0.10 |
ENST00000372680.1 |
TCEAL5 |
transcription elongation factor A (SII)-like 5 |
| chr2_-_225266711 | 0.10 |
ENST00000389874.3 |
FAM124B |
family with sequence similarity 124B |
| chr16_-_31085033 | 0.10 |
ENST00000414399.1 |
ZNF668 |
zinc finger protein 668 |
| chr2_+_24272576 | 0.10 |
ENST00000380986.4 ENST00000452109.1 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr2_-_121223697 | 0.09 |
ENST00000593290.1 |
FLJ14816 |
long intergenic non-protein coding RNA 1101 |
| chr10_-_77161650 | 0.09 |
ENST00000372524.4 |
ZNF503 |
zinc finger protein 503 |
| chr2_-_220435963 | 0.09 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr13_-_100624012 | 0.09 |
ENST00000267294.4 |
ZIC5 |
Zic family member 5 |
| chr10_+_26505179 | 0.09 |
ENST00000376261.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr11_-_61129335 | 0.09 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr15_+_31619013 | 0.09 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr17_-_36906058 | 0.09 |
ENST00000580830.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr17_+_51900239 | 0.09 |
ENST00000268919.4 |
KIF2B |
kinesin family member 2B |
| chr12_+_120740119 | 0.09 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr12_+_50366620 | 0.09 |
ENST00000315520.5 |
AQP6 |
aquaporin 6, kidney specific |
| chr2_+_185463093 | 0.09 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr11_+_61159832 | 0.08 |
ENST00000334888.5 ENST00000398979.3 |
TMEM216 |
transmembrane protein 216 |
| chr9_-_99064429 | 0.08 |
ENST00000375263.3 |
HSD17B3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr17_+_2240775 | 0.08 |
ENST00000268989.3 ENST00000426855.2 |
SGSM2 |
small G protein signaling modulator 2 |
| chr17_-_40264692 | 0.08 |
ENST00000591220.1 ENST00000251642.3 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr20_+_34742650 | 0.08 |
ENST00000373945.1 ENST00000338074.2 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr17_+_27071002 | 0.08 |
ENST00000262395.5 ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4 |
TNF receptor-associated factor 4 |
| chr1_-_153600656 | 0.08 |
ENST00000339556.4 ENST00000440685.2 |
S100A13 |
S100 calcium binding protein A13 |
| chr12_-_55028664 | 0.08 |
ENST00000547511.1 ENST00000257867.4 |
LACRT |
lacritin |
| chr17_+_4981535 | 0.08 |
ENST00000318833.3 |
ZFP3 |
ZFP3 zinc finger protein |
| chr19_-_46272462 | 0.08 |
ENST00000317578.6 |
SIX5 |
SIX homeobox 5 |
| chr3_-_99833333 | 0.08 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr12_+_108523133 | 0.08 |
ENST00000547525.1 |
WSCD2 |
WSC domain containing 2 |
| chr12_+_79258444 | 0.08 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr16_-_67969888 | 0.08 |
ENST00000574576.2 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chrX_+_135570046 | 0.08 |
ENST00000370648.3 |
BRS3 |
bombesin-like receptor 3 |
| chr9_-_100459639 | 0.08 |
ENST00000375128.4 |
XPA |
xeroderma pigmentosum, complementation group A |
| chr17_-_36891830 | 0.08 |
ENST00000578487.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr1_-_201081579 | 0.07 |
ENST00000367338.3 ENST00000362061.3 |
CACNA1S |
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr6_+_108487245 | 0.07 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr16_+_6533729 | 0.07 |
ENST00000551752.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr15_+_40531243 | 0.07 |
ENST00000558055.1 ENST00000455577.2 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr5_+_131705438 | 0.07 |
ENST00000245407.3 |
SLC22A5 |
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr2_-_128400788 | 0.07 |
ENST00000409286.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr19_+_46000479 | 0.07 |
ENST00000456399.2 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr14_+_104182061 | 0.07 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr6_+_99282570 | 0.07 |
ENST00000328345.5 |
POU3F2 |
POU class 3 homeobox 2 |
| chr10_+_23983671 | 0.07 |
ENST00000376462.1 |
KIAA1217 |
KIAA1217 |
| chr7_-_38394118 | 0.07 |
ENST00000390345.2 |
TRGV4 |
T cell receptor gamma variable 4 |
| chr14_+_63671105 | 0.07 |
ENST00000316754.3 |
RHOJ |
ras homolog family member J |
| chr10_+_104404218 | 0.07 |
ENST00000302424.7 |
TRIM8 |
tripartite motif containing 8 |
| chr1_-_49242553 | 0.07 |
ENST00000371833.3 |
BEND5 |
BEN domain containing 5 |
| chr14_+_58765103 | 0.07 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chrX_+_51075658 | 0.07 |
ENST00000356450.2 |
NUDT10 |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr2_+_210636697 | 0.07 |
ENST00000439458.1 ENST00000272845.6 |
UNC80 |
unc-80 homolog (C. elegans) |
| chr9_+_116207007 | 0.07 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr19_+_50380682 | 0.07 |
ENST00000221543.5 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr19_+_48898132 | 0.07 |
ENST00000263269.3 |
GRIN2D |
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr14_+_104182105 | 0.07 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr15_-_43876702 | 0.07 |
ENST00000348806.6 |
PPIP5K1 |
diphosphoinositol pentakisphosphate kinase 1 |
| chr9_-_130533615 | 0.07 |
ENST00000373277.4 |
SH2D3C |
SH2 domain containing 3C |
| chr3_+_154797428 | 0.07 |
ENST00000460393.1 |
MME |
membrane metallo-endopeptidase |
| chr19_-_46000251 | 0.07 |
ENST00000590526.1 ENST00000344680.4 ENST00000245923.4 |
RTN2 |
reticulon 2 |
| chr6_-_66417107 | 0.07 |
ENST00000370621.3 ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS |
eyes shut homolog (Drosophila) |
| chr6_-_33267101 | 0.07 |
ENST00000497454.1 |
RGL2 |
ral guanine nucleotide dissociation stimulator-like 2 |
| chr14_+_24605361 | 0.07 |
ENST00000206451.6 ENST00000559123.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_+_5914213 | 0.07 |
ENST00000222125.5 ENST00000452990.2 ENST00000588865.1 |
CAPS |
calcyphosine |
| chrX_-_134156502 | 0.07 |
ENST00000391440.1 |
FAM127C |
family with sequence similarity 127, member C |
| chr1_+_16083154 | 0.07 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr7_-_38403077 | 0.07 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr10_+_49917751 | 0.07 |
ENST00000325239.5 ENST00000413659.2 |
WDFY4 |
WDFY family member 4 |
| chr2_+_238875597 | 0.06 |
ENST00000272930.4 ENST00000448502.1 ENST00000416292.1 ENST00000409633.1 ENST00000414443.1 ENST00000409953.1 ENST00000409332.1 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
| chr19_+_50380917 | 0.06 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr1_-_202777535 | 0.06 |
ENST00000367264.2 |
KDM5B |
lysine (K)-specific demethylase 5B |
| chr14_+_23563952 | 0.06 |
ENST00000319074.4 |
C14orf119 |
chromosome 14 open reading frame 119 |
| chr11_-_67120974 | 0.06 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr19_+_46000506 | 0.06 |
ENST00000396737.2 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr8_+_22022653 | 0.06 |
ENST00000354870.5 ENST00000397816.3 ENST00000306349.8 |
BMP1 |
bone morphogenetic protein 1 |
| chr15_+_40531621 | 0.06 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr3_+_129247479 | 0.06 |
ENST00000296271.3 |
RHO |
rhodopsin |
| chr10_-_81205373 | 0.06 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr5_+_139493665 | 0.06 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr12_+_7060432 | 0.06 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr14_+_75348592 | 0.06 |
ENST00000334220.4 |
DLST |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr14_+_22931924 | 0.06 |
ENST00000390477.2 |
TRDC |
T cell receptor delta constant |
| chr1_+_150229554 | 0.06 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr2_+_90458201 | 0.06 |
ENST00000603238.1 |
CH17-132F21.1 |
Uncharacterized protein |
| chr7_-_100491854 | 0.06 |
ENST00000426415.1 ENST00000430554.1 ENST00000412389.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr12_+_57984965 | 0.06 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr11_+_36616044 | 0.06 |
ENST00000334307.5 ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74 |
chromosome 11 open reading frame 74 |
| chr6_+_27775899 | 0.06 |
ENST00000358739.3 |
HIST1H2AI |
histone cluster 1, H2ai |
| chr16_+_28875126 | 0.06 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr2_+_89901292 | 0.06 |
ENST00000448155.2 |
IGKV1D-39 |
immunoglobulin kappa variable 1D-39 |
| chr14_-_55878538 | 0.06 |
ENST00000247178.5 |
ATG14 |
autophagy related 14 |
| chr6_+_101846664 | 0.06 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr7_-_140178726 | 0.06 |
ENST00000480552.1 |
MKRN1 |
makorin ring finger protein 1 |
| chr8_-_98290087 | 0.05 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr11_-_63993690 | 0.05 |
ENST00000394546.2 ENST00000541278.1 |
TRPT1 |
tRNA phosphotransferase 1 |
| chr19_+_44116236 | 0.05 |
ENST00000417606.1 |
SRRM5 |
serine/arginine repetitive matrix 5 |
| chr6_+_43612750 | 0.05 |
ENST00000372165.4 ENST00000372163.4 |
RSPH9 |
radial spoke head 9 homolog (Chlamydomonas) |
| chrX_-_70288234 | 0.05 |
ENST00000276105.3 ENST00000374274.3 |
SNX12 |
sorting nexin 12 |
| chr14_+_23564700 | 0.05 |
ENST00000554203.1 |
C14orf119 |
chromosome 14 open reading frame 119 |
| chr19_+_45312347 | 0.05 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr16_+_811073 | 0.05 |
ENST00000382862.3 ENST00000563651.1 |
MSLN |
mesothelin |
| chr4_+_3443614 | 0.05 |
ENST00000382774.3 ENST00000511533.1 |
HGFAC |
HGF activator |
| chr1_-_213020991 | 0.05 |
ENST00000332912.3 |
C1orf227 |
chromosome 1 open reading frame 227 |
| chr3_-_149688502 | 0.05 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr15_-_90039805 | 0.05 |
ENST00000544600.1 ENST00000268122.4 |
RHCG |
Rh family, C glycoprotein |
| chr2_-_50201327 | 0.05 |
ENST00000412315.1 |
NRXN1 |
neurexin 1 |
| chr5_-_19988339 | 0.05 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chrX_+_54835493 | 0.05 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr17_-_17942473 | 0.05 |
ENST00000585101.1 ENST00000474627.3 ENST00000444058.1 |
ATPAF2 |
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr14_+_63671577 | 0.05 |
ENST00000555125.1 |
RHOJ |
ras homolog family member J |
| chr6_-_43021612 | 0.05 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.2 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.2 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.4 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.3 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |