Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for KLF15
Z-value: 0.16
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | KLF15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | 0.53 | 1.8e-01 | Click! |
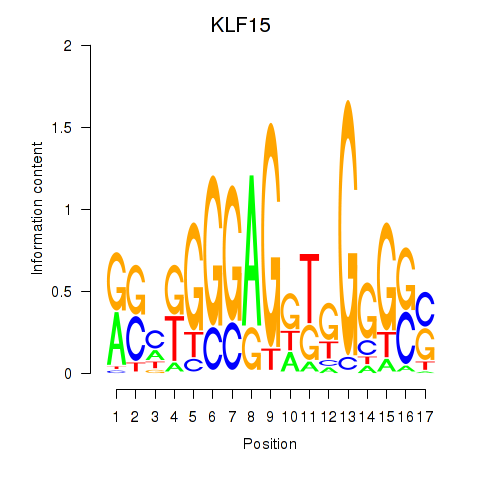
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_39640513 | 0.26 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr19_+_11457232 | 0.26 |
ENST00000587531.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr16_-_30107491 | 0.25 |
ENST00000566134.1 ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3 |
yippee-like 3 (Drosophila) |
| chr5_-_111093081 | 0.24 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr4_-_39640700 | 0.23 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr7_-_76256557 | 0.21 |
ENST00000275569.4 ENST00000310842.4 |
POMZP3 |
POM121 and ZP3 fusion |
| chr17_-_42276574 | 0.21 |
ENST00000589805.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr2_+_48757278 | 0.21 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr17_-_19290483 | 0.20 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr10_+_112631547 | 0.20 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr9_-_139891165 | 0.20 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr12_+_53443963 | 0.19 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443680 | 0.19 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr9_-_95896550 | 0.18 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr8_-_60031762 | 0.18 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr1_-_85930246 | 0.18 |
ENST00000426972.3 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr3_-_52001448 | 0.16 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr21_-_40685536 | 0.16 |
ENST00000341322.4 |
BRWD1 |
bromodomain and WD repeat domain containing 1 |
| chr6_+_32811885 | 0.16 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr17_-_19290117 | 0.16 |
ENST00000497081.2 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr14_-_89883412 | 0.16 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr12_-_58131931 | 0.15 |
ENST00000547588.1 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr14_-_80677613 | 0.15 |
ENST00000556811.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr11_-_46142615 | 0.14 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr19_-_3062465 | 0.14 |
ENST00000327141.4 |
AES |
amino-terminal enhancer of split |
| chr7_-_150675372 | 0.14 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr20_-_48099182 | 0.13 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr5_+_139027877 | 0.13 |
ENST00000302517.3 |
CXXC5 |
CXXC finger protein 5 |
| chr17_-_76921459 | 0.12 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr2_+_33172221 | 0.12 |
ENST00000354476.3 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33172012 | 0.12 |
ENST00000404816.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr19_+_41082755 | 0.12 |
ENST00000291842.5 ENST00000600733.1 |
SHKBP1 |
SH3KBP1 binding protein 1 |
| chr16_+_29819096 | 0.12 |
ENST00000568411.1 ENST00000563012.1 ENST00000562557.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_82782861 | 0.12 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr7_+_94285637 | 0.12 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr5_+_139028510 | 0.11 |
ENST00000502336.1 ENST00000520967.1 ENST00000511048.1 |
CXXC5 |
CXXC finger protein 5 |
| chr11_-_65640325 | 0.11 |
ENST00000307998.6 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
| chr17_+_73043301 | 0.11 |
ENST00000322444.6 |
KCTD2 |
potassium channel tetramerization domain containing 2 |
| chr19_+_14544099 | 0.11 |
ENST00000242783.6 ENST00000586557.1 ENST00000590097.1 |
PKN1 |
protein kinase N1 |
| chrX_-_103087136 | 0.11 |
ENST00000243298.2 |
RAB9B |
RAB9B, member RAS oncogene family |
| chr12_+_57482877 | 0.11 |
ENST00000342556.6 ENST00000357680.4 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr20_+_13976015 | 0.11 |
ENST00000217246.4 |
MACROD2 |
MACRO domain containing 2 |
| chr3_-_185826286 | 0.10 |
ENST00000537818.1 ENST00000422039.1 ENST00000434744.1 |
ETV5 |
ets variant 5 |
| chr16_-_755726 | 0.10 |
ENST00000324361.5 |
FBXL16 |
F-box and leucine-rich repeat protein 16 |
| chr14_-_65439132 | 0.10 |
ENST00000533601.2 |
RAB15 |
RAB15, member RAS oncogene family |
| chr5_+_139505520 | 0.10 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chrX_+_51927919 | 0.10 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr14_-_73493784 | 0.10 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr10_+_8096769 | 0.10 |
ENST00000346208.3 |
GATA3 |
GATA binding protein 3 |
| chr3_+_12838161 | 0.10 |
ENST00000456430.2 |
CAND2 |
cullin-associated and neddylation-dissociated 2 (putative) |
| chr12_+_9067123 | 0.09 |
ENST00000543824.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr19_-_18653781 | 0.09 |
ENST00000596558.2 ENST00000453489.2 |
FKBP8 |
FK506 binding protein 8, 38kDa |
| chr6_-_32157947 | 0.09 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr2_+_45878790 | 0.09 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr2_+_24272543 | 0.09 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr19_+_11457175 | 0.09 |
ENST00000458408.1 ENST00000586451.1 ENST00000588592.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr12_-_31744031 | 0.09 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr3_+_9851632 | 0.09 |
ENST00000426895.4 |
TTLL3 |
tubulin tyrosine ligase-like family, member 3 |
| chr11_-_46142505 | 0.09 |
ENST00000524497.1 ENST00000418153.2 |
PHF21A |
PHD finger protein 21A |
| chrX_-_1572629 | 0.09 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr11_+_1874200 | 0.09 |
ENST00000311604.3 |
LSP1 |
lymphocyte-specific protein 1 |
| chr19_-_14201776 | 0.09 |
ENST00000269724.5 |
SAMD1 |
sterile alpha motif domain containing 1 |
| chr3_-_49726486 | 0.09 |
ENST00000449682.2 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_+_43543864 | 0.09 |
ENST00000372236.4 ENST00000535400.1 |
POLH |
polymerase (DNA directed), eta |
| chr2_-_200322723 | 0.08 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr2_+_39893043 | 0.08 |
ENST00000281961.2 |
TMEM178A |
transmembrane protein 178A |
| chr7_-_28220354 | 0.08 |
ENST00000283928.5 |
JAZF1 |
JAZF zinc finger 1 |
| chr12_+_26274917 | 0.08 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr17_-_42277203 | 0.08 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr6_+_24495067 | 0.08 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr11_-_46142948 | 0.08 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr1_-_11751529 | 0.08 |
ENST00000376672.1 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr12_+_57853918 | 0.08 |
ENST00000532291.1 ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1 |
GLI family zinc finger 1 |
| chr10_-_79397479 | 0.08 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_+_24272576 | 0.08 |
ENST00000380986.4 ENST00000452109.1 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr3_+_9851904 | 0.08 |
ENST00000547186.1 ENST00000397241.1 ENST00000426827.1 |
TTLL3 |
tubulin tyrosine ligase-like family, member 3 |
| chr14_-_65438865 | 0.08 |
ENST00000267512.5 |
RAB15 |
RAB15, member RAS oncogene family |
| chr17_+_55333876 | 0.07 |
ENST00000284073.2 |
MSI2 |
musashi RNA-binding protein 2 |
| chr20_+_37353084 | 0.07 |
ENST00000217420.1 |
SLC32A1 |
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr12_+_53773944 | 0.07 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr7_+_127228399 | 0.07 |
ENST00000000233.5 ENST00000415666.1 |
ARF5 |
ADP-ribosylation factor 5 |
| chr6_+_43543942 | 0.07 |
ENST00000372226.1 ENST00000443535.1 |
POLH |
polymerase (DNA directed), eta |
| chr1_+_61547405 | 0.07 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr14_-_105262055 | 0.07 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr1_+_38273419 | 0.07 |
ENST00000468084.1 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr17_+_55334364 | 0.07 |
ENST00000322684.3 ENST00000579590.1 |
MSI2 |
musashi RNA-binding protein 2 |
| chr22_+_42949925 | 0.07 |
ENST00000327678.5 ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2 |
serine hydrolase-like 2 |
| chr1_-_19229248 | 0.07 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr10_+_60272814 | 0.07 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr3_-_39196049 | 0.07 |
ENST00000514182.1 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
| chr17_-_73178599 | 0.07 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr1_+_36273743 | 0.07 |
ENST00000373210.3 |
AGO4 |
argonaute RISC catalytic component 4 |
| chr1_+_38273818 | 0.07 |
ENST00000373042.4 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr12_+_56862301 | 0.06 |
ENST00000338146.5 |
SPRYD4 |
SPRY domain containing 4 |
| chr3_-_71774516 | 0.06 |
ENST00000425534.3 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
| chr14_-_105262016 | 0.06 |
ENST00000407796.2 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr8_+_22409193 | 0.06 |
ENST00000240123.7 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr14_+_105886275 | 0.06 |
ENST00000405646.1 |
MTA1 |
metastasis associated 1 |
| chr22_-_38484922 | 0.06 |
ENST00000428572.1 |
BAIAP2L2 |
BAI1-associated protein 2-like 2 |
| chr10_+_74927875 | 0.06 |
ENST00000242505.6 |
FAM149B1 |
family with sequence similarity 149, member B1 |
| chr9_-_84303269 | 0.06 |
ENST00000418319.1 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
| chr6_-_80247140 | 0.06 |
ENST00000392959.1 ENST00000467898.3 |
LCA5 |
Leber congenital amaurosis 5 |
| chr5_+_17217669 | 0.06 |
ENST00000322611.3 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
| chr19_-_40791211 | 0.06 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr20_-_61493115 | 0.06 |
ENST00000335351.3 ENST00000217162.5 |
TCFL5 |
transcription factor-like 5 (basic helix-loop-helix) |
| chr1_+_61548225 | 0.06 |
ENST00000371187.3 |
NFIA |
nuclear factor I/A |
| chr16_-_57513657 | 0.06 |
ENST00000566936.1 ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4 |
docking protein 4 |
| chr2_-_145277882 | 0.06 |
ENST00000465070.1 ENST00000444559.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr11_-_82782952 | 0.06 |
ENST00000534141.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr10_+_21823079 | 0.06 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr12_-_131323719 | 0.06 |
ENST00000392373.2 |
STX2 |
syntaxin 2 |
| chr8_-_145688231 | 0.06 |
ENST00000530374.1 |
CYHR1 |
cysteine/histidine-rich 1 |
| chr17_-_41984835 | 0.06 |
ENST00000520406.1 ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr10_+_8096631 | 0.06 |
ENST00000379328.3 |
GATA3 |
GATA binding protein 3 |
| chr4_-_114682936 | 0.06 |
ENST00000454265.2 ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr6_-_80247105 | 0.06 |
ENST00000369846.4 |
LCA5 |
Leber congenital amaurosis 5 |
| chr19_-_14201507 | 0.06 |
ENST00000533683.2 |
SAMD1 |
sterile alpha motif domain containing 1 |
| chr11_-_73309228 | 0.06 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr17_+_33474826 | 0.05 |
ENST00000268876.5 ENST00000433649.1 ENST00000378449.1 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr10_+_21823243 | 0.05 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr18_-_70535177 | 0.05 |
ENST00000327305.6 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr16_+_29819372 | 0.05 |
ENST00000568544.1 ENST00000569978.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_+_33474860 | 0.05 |
ENST00000394570.2 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr10_-_15210615 | 0.05 |
ENST00000378150.1 |
NMT2 |
N-myristoyltransferase 2 |
| chr14_+_24837226 | 0.05 |
ENST00000554050.1 ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr1_+_97187318 | 0.05 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr12_-_48213568 | 0.05 |
ENST00000080059.7 ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7 |
histone deacetylase 7 |
| chr5_+_110559784 | 0.05 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr20_+_34894247 | 0.05 |
ENST00000373913.3 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr4_-_114682597 | 0.05 |
ENST00000394524.3 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr21_-_27107283 | 0.05 |
ENST00000284971.3 ENST00000400099.1 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr17_-_74099772 | 0.05 |
ENST00000411744.2 ENST00000332065.5 |
EXOC7 |
exocyst complex component 7 |
| chr15_+_57668695 | 0.05 |
ENST00000281282.5 |
CGNL1 |
cingulin-like 1 |
| chr10_+_35415978 | 0.05 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr7_+_66093851 | 0.05 |
ENST00000275532.3 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
| chr1_+_206680879 | 0.05 |
ENST00000355294.4 ENST00000367117.3 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
| chr19_-_46272462 | 0.05 |
ENST00000317578.6 |
SIX5 |
SIX homeobox 5 |
| chr1_+_51701924 | 0.05 |
ENST00000242719.3 |
RNF11 |
ring finger protein 11 |
| chr18_+_3449821 | 0.05 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr9_+_125703282 | 0.05 |
ENST00000373647.4 ENST00000402311.1 |
RABGAP1 |
RAB GTPase activating protein 1 |
| chr1_+_38273988 | 0.05 |
ENST00000446260.2 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr14_+_105886150 | 0.05 |
ENST00000331320.7 ENST00000406191.1 |
MTA1 |
metastasis associated 1 |
| chr11_+_64058758 | 0.05 |
ENST00000538767.1 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr15_+_31619013 | 0.05 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr5_+_131593364 | 0.05 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr15_-_45480147 | 0.05 |
ENST00000560734.1 |
SHF |
Src homology 2 domain containing F |
| chr17_-_73043046 | 0.05 |
ENST00000301587.4 ENST00000344546.4 |
ATP5H |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d |
| chr11_+_64058820 | 0.05 |
ENST00000422670.2 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr10_+_35416090 | 0.05 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr19_+_35759824 | 0.05 |
ENST00000343550.5 |
USF2 |
upstream transcription factor 2, c-fos interacting |
| chr10_+_35416223 | 0.04 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr19_+_3359561 | 0.04 |
ENST00000589123.1 ENST00000346156.5 ENST00000395111.3 ENST00000586919.1 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr4_+_71768043 | 0.04 |
ENST00000502869.1 ENST00000309395.2 ENST00000396051.2 |
MOB1B |
MOB kinase activator 1B |
| chr14_+_24867992 | 0.04 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr21_-_27107344 | 0.04 |
ENST00000457143.2 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr7_+_151038785 | 0.04 |
ENST00000413040.2 ENST00000568733.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr18_-_74207146 | 0.04 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr2_-_172017343 | 0.04 |
ENST00000431350.2 ENST00000360843.3 |
TLK1 |
tousled-like kinase 1 |
| chr8_-_116681221 | 0.04 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr2_+_242254507 | 0.04 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chr16_+_29819446 | 0.04 |
ENST00000568282.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr12_-_50101165 | 0.04 |
ENST00000352151.5 ENST00000335154.5 ENST00000293590.5 |
FMNL3 |
formin-like 3 |
| chr14_-_67982146 | 0.04 |
ENST00000557779.1 ENST00000557006.1 |
TMEM229B |
transmembrane protein 229B |
| chr3_-_105587879 | 0.04 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr17_+_75277492 | 0.04 |
ENST00000427177.1 ENST00000591198.1 |
SEPT9 |
septin 9 |
| chr11_+_45918092 | 0.04 |
ENST00000395629.2 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_-_88108192 | 0.04 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr17_-_74099795 | 0.04 |
ENST00000406660.3 ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7 |
exocyst complex component 7 |
| chr19_-_4124079 | 0.04 |
ENST00000394867.4 ENST00000262948.5 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
| chr16_-_11485922 | 0.04 |
ENST00000599216.1 |
CTD-3088G3.8 |
Protein LOC388210 |
| chr7_-_72971934 | 0.04 |
ENST00000411832.1 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr7_-_100493482 | 0.04 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr21_+_30671189 | 0.04 |
ENST00000286800.3 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr3_-_149688502 | 0.04 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr11_-_8954491 | 0.04 |
ENST00000526227.1 ENST00000525780.1 ENST00000326053.5 |
C11orf16 |
chromosome 11 open reading frame 16 |
| chr8_+_55370487 | 0.04 |
ENST00000297316.4 |
SOX17 |
SRY (sex determining region Y)-box 17 |
| chr11_-_72496976 | 0.04 |
ENST00000539138.1 ENST00000542989.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr1_-_153895377 | 0.04 |
ENST00000368655.4 |
GATAD2B |
GATA zinc finger domain containing 2B |
| chr3_-_49449350 | 0.04 |
ENST00000454011.2 ENST00000445425.1 ENST00000422781.1 |
RHOA |
ras homolog family member A |
| chr10_+_28822417 | 0.04 |
ENST00000428935.1 ENST00000420266.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr17_-_79633590 | 0.04 |
ENST00000374741.3 ENST00000571503.1 |
OXLD1 |
oxidoreductase-like domain containing 1 |
| chr13_-_49107303 | 0.04 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr5_+_110559603 | 0.04 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr19_-_14049184 | 0.04 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr19_+_852291 | 0.04 |
ENST00000263621.1 |
ELANE |
elastase, neutrophil expressed |
| chr9_-_14314066 | 0.04 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr12_+_56137064 | 0.04 |
ENST00000257868.5 ENST00000546799.1 |
GDF11 |
growth differentiation factor 11 |
| chr6_-_74230741 | 0.04 |
ENST00000316292.9 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr17_+_48911942 | 0.04 |
ENST00000426127.1 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr12_+_9067327 | 0.04 |
ENST00000433083.2 ENST00000544916.1 ENST00000544539.1 ENST00000539063.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr2_+_139259324 | 0.04 |
ENST00000280098.4 |
SPOPL |
speckle-type POZ protein-like |
| chr6_-_34664612 | 0.04 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr18_-_21977748 | 0.04 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr10_-_25012115 | 0.04 |
ENST00000446003.1 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr8_+_38758737 | 0.04 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr5_+_42423872 | 0.04 |
ENST00000230882.4 ENST00000357703.3 |
GHR |
growth hormone receptor |
| chr21_-_27107198 | 0.04 |
ENST00000400094.1 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr10_-_133795416 | 0.04 |
ENST00000540159.1 ENST00000368636.4 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr10_+_181418 | 0.04 |
ENST00000403354.1 ENST00000381607.4 ENST00000402736.1 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr1_+_43148625 | 0.04 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036446 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0061289 | pro-T cell differentiation(GO:0002572) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0006540 | acetate metabolic process(GO:0006083) glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.0 | GO:0061010 | cardiac cell fate determination(GO:0060913) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |