Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for KLF3
Z-value: 0.27
Transcription factors associated with KLF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
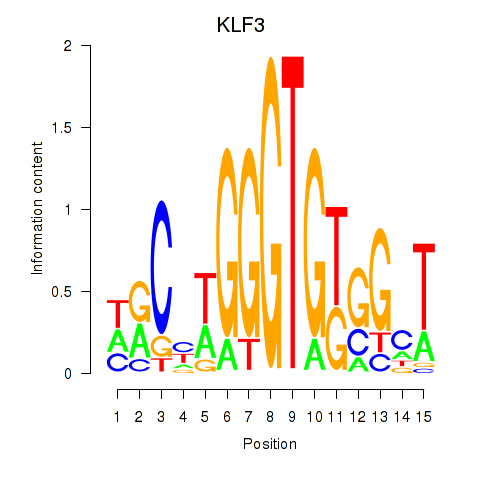
KLF3
|
ENSG00000109787.8 | KLF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF3 | hg19_v2_chr4_+_38665810_38665827 | -0.69 | 5.6e-02 | Click! |
Activity profile of KLF3 motif
Sorted Z-values of KLF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_102104980 | 0.48 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr13_+_102104952 | 0.37 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chrX_-_152486108 | 0.34 |
ENST00000356661.5 |
MAGEA1 |
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
| chr5_+_75699149 | 0.30 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr1_-_57045228 | 0.28 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr3_+_49449636 | 0.27 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr4_-_89619386 | 0.21 |
ENST00000323061.5 |
NAP1L5 |
nucleosome assembly protein 1-like 5 |
| chr20_+_36531499 | 0.20 |
ENST00000373458.3 ENST00000373461.4 ENST00000373459.4 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
| chr14_-_106406090 | 0.20 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chr10_+_94451574 | 0.18 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr16_+_1578674 | 0.17 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr17_-_76870126 | 0.17 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr8_-_13134045 | 0.16 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr2_+_217498105 | 0.16 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr22_-_32341336 | 0.16 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr5_-_150521192 | 0.16 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr17_+_38599693 | 0.16 |
ENST00000542955.1 ENST00000269593.4 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
| chr1_-_33168336 | 0.15 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr12_-_56236690 | 0.14 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr3_+_54156664 | 0.14 |
ENST00000474759.1 ENST00000288197.5 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr3_+_54156570 | 0.14 |
ENST00000415676.2 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr7_-_12443501 | 0.13 |
ENST00000275358.3 |
VWDE |
von Willebrand factor D and EGF domains |
| chr20_+_3052264 | 0.13 |
ENST00000217386.2 |
OXT |
oxytocin/neurophysin I prepropeptide |
| chr11_-_111783919 | 0.13 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr17_-_48278983 | 0.13 |
ENST00000225964.5 |
COL1A1 |
collagen, type I, alpha 1 |
| chr22_+_35776828 | 0.12 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr3_+_51976338 | 0.12 |
ENST00000417220.2 ENST00000431474.1 ENST00000398755.3 |
PARP3 |
poly (ADP-ribose) polymerase family, member 3 |
| chr19_-_14016877 | 0.12 |
ENST00000454313.1 ENST00000591586.1 ENST00000346736.2 |
C19orf57 |
chromosome 19 open reading frame 57 |
| chr12_-_54121261 | 0.12 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr14_+_103566481 | 0.12 |
ENST00000380069.3 |
EXOC3L4 |
exocyst complex component 3-like 4 |
| chr14_+_22458631 | 0.12 |
ENST00000390444.1 |
TRAV16 |
T cell receptor alpha variable 16 |
| chr11_-_111783595 | 0.12 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr7_+_29603394 | 0.11 |
ENST00000319694.2 |
PRR15 |
proline rich 15 |
| chr17_-_19290483 | 0.11 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr19_-_3025614 | 0.11 |
ENST00000447365.2 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr12_+_7169887 | 0.11 |
ENST00000542978.1 |
C1S |
complement component 1, s subcomponent |
| chr4_+_26585538 | 0.11 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr2_+_210288760 | 0.11 |
ENST00000199940.6 |
MAP2 |
microtubule-associated protein 2 |
| chr10_-_102089729 | 0.11 |
ENST00000465680.2 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr1_-_60539405 | 0.11 |
ENST00000450089.2 |
C1orf87 |
chromosome 1 open reading frame 87 |
| chr1_-_60539422 | 0.11 |
ENST00000371201.3 |
C1orf87 |
chromosome 1 open reading frame 87 |
| chr3_+_106959530 | 0.11 |
ENST00000466734.1 ENST00000463143.1 ENST00000490441.1 |
LINC00883 |
long intergenic non-protein coding RNA 883 |
| chr2_+_71295733 | 0.11 |
ENST00000443938.2 ENST00000244204.6 |
NAGK |
N-acetylglucosamine kinase |
| chr2_+_172378757 | 0.10 |
ENST00000409484.1 ENST00000321348.4 ENST00000375252.3 |
CYBRD1 |
cytochrome b reductase 1 |
| chr11_+_75428857 | 0.10 |
ENST00000198801.5 |
MOGAT2 |
monoacylglycerol O-acyltransferase 2 |
| chrX_-_13956737 | 0.10 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chr2_+_71295717 | 0.10 |
ENST00000418807.3 ENST00000443872.2 |
NAGK |
N-acetylglucosamine kinase |
| chr3_+_11196206 | 0.10 |
ENST00000431010.2 |
HRH1 |
histamine receptor H1 |
| chr6_-_35109080 | 0.10 |
ENST00000486638.1 ENST00000505400.1 ENST00000412155.2 ENST00000373979.2 ENST00000507706.1 ENST00000444780.2 ENST00000492680.2 |
TCP11 |
t-complex 11, testis-specific |
| chr2_+_169923577 | 0.10 |
ENST00000432060.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr12_-_54121212 | 0.10 |
ENST00000548263.1 ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr2_+_169923504 | 0.10 |
ENST00000357546.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr16_+_85061367 | 0.10 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr3_+_141144963 | 0.10 |
ENST00000510726.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr20_+_327668 | 0.10 |
ENST00000382291.3 ENST00000609504.1 ENST00000382285.2 |
NRSN2 |
neurensin 2 |
| chr16_+_66400533 | 0.10 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr6_+_30524663 | 0.09 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr7_+_33169142 | 0.09 |
ENST00000242067.6 ENST00000350941.3 ENST00000396127.2 ENST00000355070.2 ENST00000354265.4 ENST00000425508.2 |
BBS9 |
Bardet-Biedl syndrome 9 |
| chr17_+_34312621 | 0.09 |
ENST00000591669.1 |
CTB-186H2.3 |
Uncharacterized protein |
| chr2_+_242811874 | 0.09 |
ENST00000343216.3 |
CXXC11 |
CXXC finger protein 11 |
| chr12_-_81331460 | 0.09 |
ENST00000549417.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chr9_+_127539481 | 0.09 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr19_+_41860047 | 0.09 |
ENST00000604123.1 |
TMEM91 |
transmembrane protein 91 |
| chr17_-_15496722 | 0.09 |
ENST00000472534.1 |
CDRT1 |
CMT1A duplicated region transcript 1 |
| chr3_-_49449521 | 0.09 |
ENST00000431929.1 ENST00000418115.1 |
RHOA |
ras homolog family member A |
| chr5_+_133842243 | 0.09 |
ENST00000515627.2 |
AC005355.2 |
AC005355.2 |
| chr19_-_14064114 | 0.09 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chrX_-_101397433 | 0.09 |
ENST00000372774.3 |
TCEAL6 |
transcription elongation factor A (SII)-like 6 |
| chr7_-_150974494 | 0.09 |
ENST00000392811.2 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr3_+_20081515 | 0.09 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr1_+_171227069 | 0.09 |
ENST00000354841.4 |
FMO1 |
flavin containing monooxygenase 1 |
| chr16_+_67207838 | 0.09 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr7_-_102184083 | 0.09 |
ENST00000379357.5 |
POLR2J3 |
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr16_+_67207872 | 0.09 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr6_+_30525051 | 0.08 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr3_+_133118839 | 0.08 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr9_-_34665983 | 0.08 |
ENST00000416454.1 ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5 |
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr9_-_130637244 | 0.08 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr7_-_100491854 | 0.08 |
ENST00000426415.1 ENST00000430554.1 ENST00000412389.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr11_-_65793948 | 0.08 |
ENST00000312106.5 |
CATSPER1 |
cation channel, sperm associated 1 |
| chr15_+_41523417 | 0.08 |
ENST00000560397.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr1_+_114471809 | 0.08 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr2_-_79315112 | 0.08 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr15_-_20193370 | 0.08 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_-_219925189 | 0.08 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr12_-_96390108 | 0.08 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr21_+_38445539 | 0.08 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr6_+_46661575 | 0.08 |
ENST00000450697.1 |
TDRD6 |
tudor domain containing 6 |
| chr16_+_89696692 | 0.08 |
ENST00000261615.4 |
DPEP1 |
dipeptidase 1 (renal) |
| chr5_+_134240800 | 0.08 |
ENST00000512783.1 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr2_+_48757278 | 0.07 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr1_+_38022572 | 0.07 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr1_+_17944832 | 0.07 |
ENST00000167825.4 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr20_+_2795626 | 0.07 |
ENST00000603872.1 ENST00000380589.4 |
C20orf141 |
chromosome 20 open reading frame 141 |
| chr2_-_85637459 | 0.07 |
ENST00000409921.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr16_+_29973351 | 0.07 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr6_+_41010293 | 0.07 |
ENST00000373161.1 ENST00000373158.2 ENST00000470917.1 |
TSPO2 |
translocator protein 2 |
| chr1_-_154461642 | 0.07 |
ENST00000555188.1 |
SHE |
Src homology 2 domain containing E |
| chrX_+_52511925 | 0.07 |
ENST00000375588.1 |
XAGE1C |
X antigen family, member 1C |
| chr19_+_41107249 | 0.07 |
ENST00000396819.3 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chrX_-_52260199 | 0.07 |
ENST00000375600.1 |
XAGE1A |
X antigen family, member 1A |
| chr12_-_81331697 | 0.07 |
ENST00000552864.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chr16_+_66878814 | 0.07 |
ENST00000394069.3 |
CA7 |
carbonic anhydrase VII |
| chr16_+_31483451 | 0.07 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr22_-_24322660 | 0.07 |
ENST00000404092.1 |
DDT |
D-dopachrome tautomerase |
| chr1_+_210406121 | 0.07 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr7_-_102283238 | 0.07 |
ENST00000340457.8 |
UPK3BL |
uroplakin 3B-like |
| chr16_+_31483374 | 0.07 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr1_-_238054094 | 0.07 |
ENST00000366570.4 |
ZP4 |
zona pellucida glycoprotein 4 |
| chr11_-_62313090 | 0.07 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chrX_+_153770421 | 0.07 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr3_-_49449350 | 0.06 |
ENST00000454011.2 ENST00000445425.1 ENST00000422781.1 |
RHOA |
ras homolog family member A |
| chr6_-_35109145 | 0.06 |
ENST00000373974.4 ENST00000244645.3 |
TCP11 |
t-complex 11, testis-specific |
| chr17_+_33914460 | 0.06 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_54468974 | 0.06 |
ENST00000381375.2 ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B |
cell division cycle 20B |
| chr19_-_10464570 | 0.06 |
ENST00000529739.1 |
TYK2 |
tyrosine kinase 2 |
| chr15_+_63334831 | 0.06 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr16_-_1020849 | 0.06 |
ENST00000568897.1 |
LMF1 |
lipase maturation factor 1 |
| chrX_+_102631844 | 0.06 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chrX_-_52533139 | 0.06 |
ENST00000374959.3 |
XAGE1D |
X antigen family, member 1D |
| chrX_-_71526813 | 0.06 |
ENST00000246139.5 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr4_-_82136114 | 0.06 |
ENST00000395578.1 ENST00000418486.2 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
| chr22_-_20461786 | 0.06 |
ENST00000426804.1 |
RIMBP3 |
RIMS binding protein 3 |
| chr10_+_104614008 | 0.06 |
ENST00000369883.3 |
C10orf32 |
chromosome 10 open reading frame 32 |
| chr4_+_2845547 | 0.06 |
ENST00000264758.7 ENST00000446856.1 ENST00000398125.1 |
ADD1 |
adducin 1 (alpha) |
| chr1_+_17944806 | 0.06 |
ENST00000375408.3 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chrX_-_71526999 | 0.06 |
ENST00000453707.2 ENST00000373619.3 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr19_-_58238958 | 0.06 |
ENST00000596939.1 ENST00000335820.3 ENST00000317398.6 |
ZNF671 |
zinc finger protein 671 |
| chr17_+_21279509 | 0.06 |
ENST00000583088.1 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr21_-_47738112 | 0.06 |
ENST00000417060.1 |
C21orf58 |
chromosome 21 open reading frame 58 |
| chr19_-_40023450 | 0.06 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr22_+_21737663 | 0.06 |
ENST00000434111.1 |
RIMBP3B |
RIMS binding protein 3B |
| chrX_-_62974941 | 0.06 |
ENST00000374872.1 ENST00000253401.6 ENST00000374870.4 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chrX_-_65858865 | 0.06 |
ENST00000374719.3 ENST00000450752.1 ENST00000451436.2 |
EDA2R |
ectodysplasin A2 receptor |
| chr11_+_1874200 | 0.06 |
ENST00000311604.3 |
LSP1 |
lymphocyte-specific protein 1 |
| chr19_-_10450328 | 0.06 |
ENST00000160262.5 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr19_-_4455290 | 0.06 |
ENST00000394765.3 ENST00000592515.1 |
UBXN6 |
UBX domain protein 6 |
| chr19_-_12945362 | 0.06 |
ENST00000590404.1 ENST00000592204.1 |
RTBDN |
retbindin |
| chr19_-_10450287 | 0.06 |
ENST00000589261.1 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr8_-_70745575 | 0.06 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr20_+_39765581 | 0.06 |
ENST00000244007.3 |
PLCG1 |
phospholipase C, gamma 1 |
| chr7_+_44788430 | 0.05 |
ENST00000457123.1 ENST00000309315.4 |
ZMIZ2 |
zinc finger, MIZ-type containing 2 |
| chr11_-_414948 | 0.05 |
ENST00000530494.1 ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr12_+_44152740 | 0.05 |
ENST00000440781.2 ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4 |
interleukin-1 receptor-associated kinase 4 |
| chr11_+_65407331 | 0.05 |
ENST00000527525.1 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr10_+_115438920 | 0.05 |
ENST00000429617.1 ENST00000369331.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr3_+_42695176 | 0.05 |
ENST00000232974.6 ENST00000457842.3 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
| chr5_-_176433693 | 0.05 |
ENST00000507513.1 ENST00000511320.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr19_-_40931891 | 0.05 |
ENST00000357949.4 |
SERTAD1 |
SERTA domain containing 1 |
| chr17_-_19619917 | 0.05 |
ENST00000325411.5 ENST00000350657.5 ENST00000433844.2 |
SLC47A2 |
solute carrier family 47 (multidrug and toxin extrusion), member 2 |
| chr6_-_31651817 | 0.05 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr19_-_40971667 | 0.05 |
ENST00000263368.4 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr4_-_681114 | 0.05 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chrX_-_52546033 | 0.05 |
ENST00000375567.3 |
XAGE1E |
X antigen family, member 1E |
| chr17_-_8198636 | 0.05 |
ENST00000577745.1 ENST00000579192.1 ENST00000396278.1 |
SLC25A35 |
solute carrier family 25, member 35 |
| chr3_-_127542051 | 0.05 |
ENST00000398104.1 |
MGLL |
monoglyceride lipase |
| chr19_-_40971643 | 0.05 |
ENST00000595483.1 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr12_+_7013897 | 0.05 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr17_+_1633755 | 0.05 |
ENST00000545662.1 |
WDR81 |
WD repeat domain 81 |
| chr11_-_124670550 | 0.05 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr17_-_39623681 | 0.05 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr15_-_101084446 | 0.05 |
ENST00000538112.2 ENST00000559639.1 ENST00000558884.2 |
CERS3 |
ceramide synthase 3 |
| chr11_-_6426635 | 0.05 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_67205538 | 0.05 |
ENST00000326294.3 |
PTPRCAP |
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr1_+_180165672 | 0.05 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr17_-_36413133 | 0.05 |
ENST00000523089.1 ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2 |
TBC1 domain family member 3 |
| chr2_+_220143989 | 0.05 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_23695680 | 0.05 |
ENST00000454117.1 ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213 |
chromosome 1 open reading frame 213 |
| chr3_+_141144954 | 0.05 |
ENST00000441582.2 ENST00000321464.5 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chrX_-_65253506 | 0.05 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr15_-_101084547 | 0.05 |
ENST00000394113.1 |
CERS3 |
ceramide synthase 3 |
| chr5_-_176433582 | 0.05 |
ENST00000506128.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr22_+_38071615 | 0.05 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr5_-_176433350 | 0.05 |
ENST00000377227.4 ENST00000377219.2 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr8_-_145060593 | 0.05 |
ENST00000313059.5 ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10 |
poly (ADP-ribose) polymerase family, member 10 |
| chr3_-_50383096 | 0.05 |
ENST00000442887.1 ENST00000360165.3 |
ZMYND10 |
zinc finger, MYND-type containing 10 |
| chr10_+_89117425 | 0.05 |
ENST00000381697.2 ENST00000412718.1 |
NUTM2D |
NUT family member 2D |
| chr2_-_85636928 | 0.05 |
ENST00000449030.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr11_+_8932715 | 0.05 |
ENST00000529876.1 ENST00000525005.1 ENST00000524577.1 ENST00000534506.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr3_-_10052763 | 0.05 |
ENST00000383808.2 ENST00000426698.1 ENST00000470827.2 |
AC022007.5 EMC3 |
AC022007.5 ER membrane protein complex subunit 3 |
| chr15_+_40643227 | 0.05 |
ENST00000448599.2 |
PHGR1 |
proline/histidine/glycine-rich 1 |
| chr22_+_31523734 | 0.05 |
ENST00000402238.1 ENST00000404453.1 ENST00000401755.1 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr2_+_131513008 | 0.05 |
ENST00000321420.4 |
AMER3 |
APC membrane recruitment protein 3 |
| chr2_+_54951679 | 0.05 |
ENST00000356458.6 |
EML6 |
echinoderm microtubule associated protein like 6 |
| chr17_+_33914276 | 0.05 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chrX_-_65859096 | 0.05 |
ENST00000456230.2 |
EDA2R |
ectodysplasin A2 receptor |
| chr16_+_28943260 | 0.05 |
ENST00000538922.1 ENST00000324662.3 ENST00000567541.1 |
CD19 |
CD19 molecule |
| chr19_-_7936344 | 0.05 |
ENST00000599142.1 |
CTD-3193O13.9 |
Protein FLJ22184 |
| chr5_+_173315283 | 0.05 |
ENST00000265085.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr16_-_3149278 | 0.04 |
ENST00000575108.1 ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10 |
zinc finger and SCAN domain containing 10 |
| chr5_-_138775177 | 0.04 |
ENST00000302060.5 |
DNAJC18 |
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr17_+_36873677 | 0.04 |
ENST00000471200.1 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr10_+_81462983 | 0.04 |
ENST00000448135.1 ENST00000429828.1 ENST00000372321.1 |
NUTM2B |
NUT family member 2B |
| chr17_-_61781750 | 0.04 |
ENST00000582026.1 |
STRADA |
STE20-related kinase adaptor alpha |
| chr3_-_139199565 | 0.04 |
ENST00000511956.1 |
RBP2 |
retinol binding protein 2, cellular |
| chr15_-_41522889 | 0.04 |
ENST00000458580.2 ENST00000314992.5 ENST00000558396.1 |
EXD1 |
exonuclease 3'-5' domain containing 1 |
| chr15_-_90234006 | 0.04 |
ENST00000300056.3 ENST00000559170.1 |
PEX11A |
peroxisomal biogenesis factor 11 alpha |
| chr20_-_17641097 | 0.04 |
ENST00000246043.4 |
RRBP1 |
ribosome binding protein 1 |
| chr17_-_32690239 | 0.04 |
ENST00000225842.3 |
CCL1 |
chemokine (C-C motif) ligand 1 |
| chr11_-_2906979 | 0.04 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.2 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.2 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:2000360 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) negative regulation of protein import into nucleus, translocation(GO:0033159) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.0 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.0 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |