Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for KLF7
Z-value: 0.57
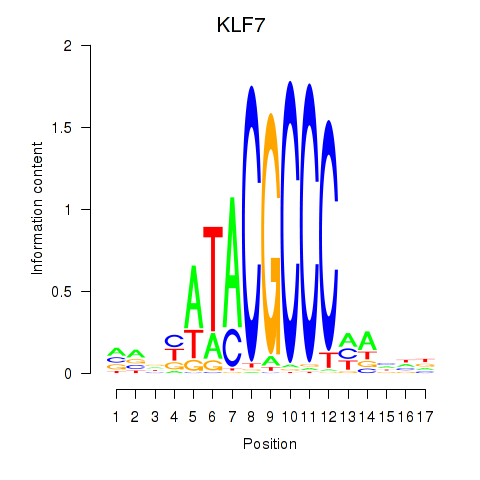
Transcription factors associated with KLF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF7
|
ENSG00000118263.10 | KLF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF7 | hg19_v2_chr2_-_208031943_208031991 | -0.78 | 2.1e-02 | Click! |
Activity profile of KLF7 motif
Sorted Z-values of KLF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_19281203 | 0.26 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr1_-_161102367 | 0.19 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr2_-_188430478 | 0.16 |
ENST00000421427.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_+_141348721 | 0.15 |
ENST00000507163.1 ENST00000394519.1 |
RNF14 |
ring finger protein 14 |
| chr14_+_29234870 | 0.12 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr5_+_173315283 | 0.12 |
ENST00000265085.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chrX_-_92928557 | 0.12 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr2_+_97779233 | 0.10 |
ENST00000461153.2 ENST00000420699.2 |
ANKRD36 |
ankyrin repeat domain 36 |
| chr3_+_186353756 | 0.09 |
ENST00000431018.1 ENST00000450521.1 ENST00000539949.1 |
FETUB |
fetuin B |
| chr3_+_105085734 | 0.09 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr5_+_141348598 | 0.09 |
ENST00000394520.2 ENST00000347642.3 |
RNF14 |
ring finger protein 14 |
| chr5_+_176560007 | 0.08 |
ENST00000510954.1 ENST00000354179.4 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr15_+_92937058 | 0.08 |
ENST00000268164.3 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr20_-_2821756 | 0.08 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr6_-_30585009 | 0.08 |
ENST00000376511.2 |
PPP1R10 |
protein phosphatase 1, regulatory subunit 10 |
| chr19_-_40931891 | 0.07 |
ENST00000357949.4 |
SERTAD1 |
SERTA domain containing 1 |
| chr3_-_105588231 | 0.07 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr1_-_161102421 | 0.07 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr15_+_96873921 | 0.07 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr20_-_2821271 | 0.07 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chrX_+_102631844 | 0.07 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr1_-_19229248 | 0.07 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_19229014 | 0.06 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr12_-_49582593 | 0.06 |
ENST00000295766.5 |
TUBA1A |
tubulin, alpha 1a |
| chr19_-_39390440 | 0.05 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr19_-_39390350 | 0.05 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr2_+_67624430 | 0.05 |
ENST00000272342.5 |
ETAA1 |
Ewing tumor-associated antigen 1 |
| chr8_-_6837602 | 0.04 |
ENST00000382692.2 |
DEFA1 |
defensin, alpha 1 |
| chr16_+_20499024 | 0.04 |
ENST00000593357.1 |
AC137056.1 |
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr3_-_105587879 | 0.04 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr8_-_6875778 | 0.04 |
ENST00000535841.1 ENST00000327857.2 |
DEFA1B DEFA3 |
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr20_+_2821340 | 0.03 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr8_+_17104539 | 0.03 |
ENST00000521829.1 ENST00000521005.1 |
VPS37A |
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr6_+_31865552 | 0.03 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr3_+_178866199 | 0.03 |
ENST00000263967.3 |
PIK3CA |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr19_+_39390320 | 0.03 |
ENST00000576510.1 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr16_+_81772633 | 0.03 |
ENST00000566191.1 ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1 PLCG2 |
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr6_-_109703600 | 0.02 |
ENST00000512821.1 |
CD164 |
CD164 molecule, sialomucin |
| chr8_-_17104356 | 0.02 |
ENST00000361272.4 ENST00000523917.1 |
CNOT7 |
CCR4-NOT transcription complex, subunit 7 |
| chr15_+_92937144 | 0.02 |
ENST00000539113.1 ENST00000555434.1 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr2_-_74710078 | 0.01 |
ENST00000290418.4 |
CCDC142 |
coiled-coil domain containing 142 |
| chr2_+_74710194 | 0.01 |
ENST00000410003.1 ENST00000442235.2 ENST00000233623.5 |
TTC31 |
tetratricopeptide repeat domain 31 |
| chr1_+_28157273 | 0.01 |
ENST00000311772.5 ENST00000236412.7 ENST00000373931.4 |
PPP1R8 |
protein phosphatase 1, regulatory subunit 8 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.0 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |